Anaphylactic Skin Reaction Detection during Chemotherapy powered by Nvidia
DISCLAIMER: This application is used for demonstrative and illustrative purposes only and does not constitute an offering that has gone through regulatory review. It is not intended to serve as a medical application. There is no representation as to the accuracy of the output of this application and it is presented without warranty.
WARNING: Some medical images are used to encapsulate the problem. Refrain if you are sensitive to that content.
Introduction:Nowadays we have serious problems in healthcare. Despite the fact that many of them have already been solved with technology, many times the industry is reluctant to change and to adopt new technologies.
Cancer patients, for example, may have to be treated with chemotherapy continuously, however this process is not simple or pleasant. The worst part of this, is the possibility of being allergic to chemotherapy. Approximately 17% of patients who are treated with chemotherapy have an allergic reaction to it, which can trigger symptoms that go from a simple blush to death. This is a very studied phenomenon which have several symptoms and phases.
One of the main symptoms (1) before severe anaphylaxis in this type of treatment is face flushing or blushing, after that there may be symptoms such as these:
If the first symptoms, which are flushing, blushing and facial redness can be detected faster, we can avoid most of the more severe ones by administering treatment promptly.
This problem is quite severe and happens even in top of the line hospitals (2). Personally I worked in one of these, and to my surprise these kinds of reactions happen quite often and I think that AI and CV technologies have the capacity to tend to this problem faster and directly so medical professionals can have a faster reaction to it.
The type of technologies to help against this problem do not exist. The first response in hospital environments happens when vital signs monitors begin to detect abnormalities in breathing. And in the event that the patient connected to one is not allotted, then the response will take place (maybe a little too late) when the patient calls the nurse once the victim begins to feel bad.
There are some technologies that serve as prevention against allergic reactions such as:
Autoinjector
- https://wyss.harvard.edu/technology/project-abbie/
- https://snacksafely.com/2018/10/project-abbie-promises-wearable-auto-injector-with-automatic-anaphylaxis-detection-and-activation/
Nevertheless these kinds of solutions are a last resource, and are not recommended.
This solution would be very useful in this type of environments, since it is a problem that nobody is solving and greatly affects the patient's well-being. Such a reaction can easily lead to complications, and if treatment is provided before the onset of severe symptoms, that may even save the patient's life.
Intent
For these reasons I will build a CV system that analyzes the patient's face in real time and is able to determine if the person is having or starting to have an allergic reaction to chemotherapy drugs. And in turn, can make an emergency call to nurses. In the experience of several nurses at the hospital, after the first symptom of the redness, the patient begins to have very aggressive allergic reactions and its very hard for the nurses to perceive this.
The solution will analyze through a 1080p camera, real-time images of the patient. Once the image capture is done, it will analyze it using a model. If the patient is presenting an allergic reaction, or if he had an allergic reaction, a notification will be sent via AWSIoT to a progressive Web App integrated with AWS-SDK. The web page will be designed with ReactJS.
The system will have the following characteristics and features:
- Real-time analysis of the patient's anaphylaxis status.
- Notifications whenever an allergic reaction occurs.
- Cloud integration of data analytics.
The model to be used will be a TensorFlow model, which I will train with several images of patients with an anaphylactic reaction (face redness) and healthy patients.
This type of models can be used with Keras (TensorFlow 2.0 submodule) and OpenCV to perform image preprocessing, training and analysis (predictions with the model). The model’s training will be done in a Jupyter notebook and that will take part on the NVIDIA Jetson Nano. This, in order to be able to generate more and more precise models, using new images that the system will collect over time and in turn, retrain the existing model. With that I hope to generate a model that learns as it sees more patients.
Connection Diagram:This is the connection diagram of the system:
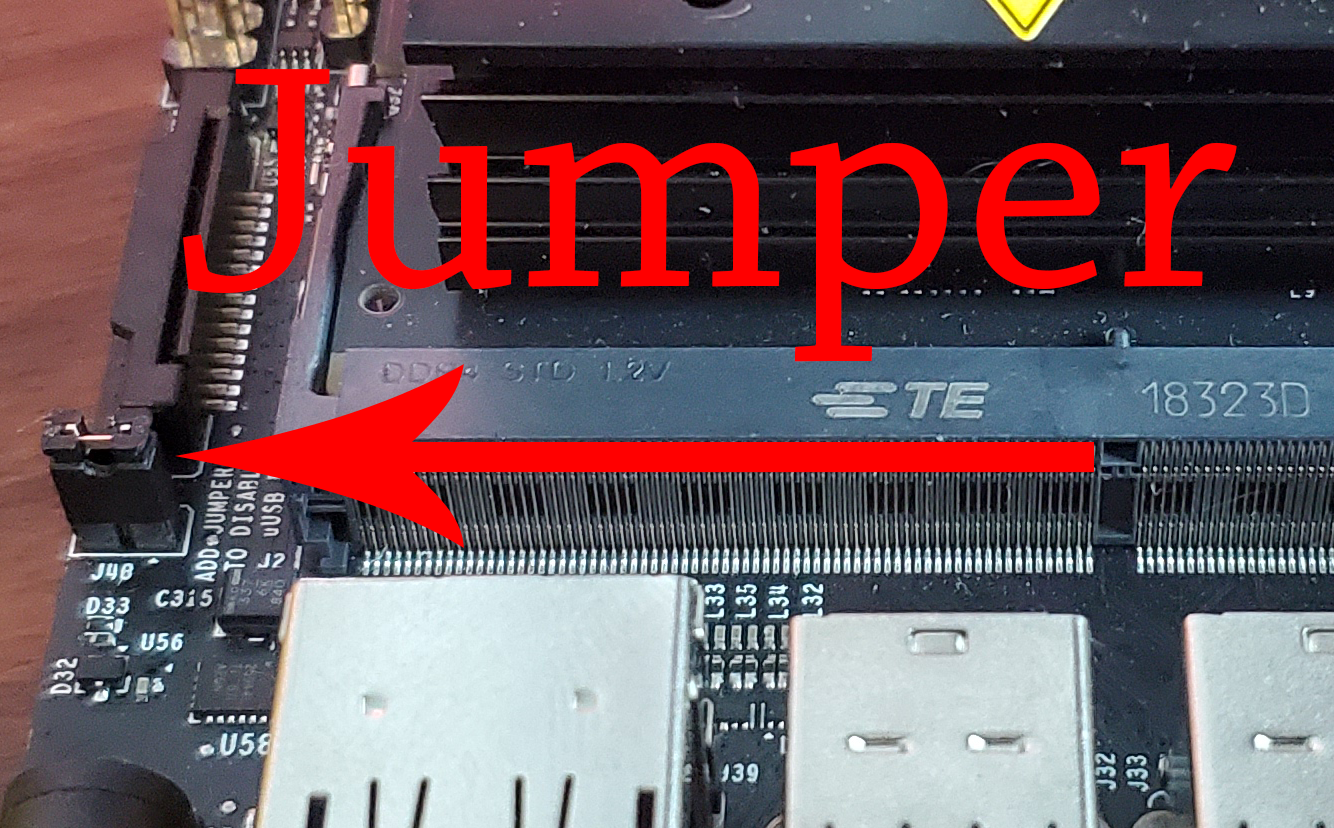
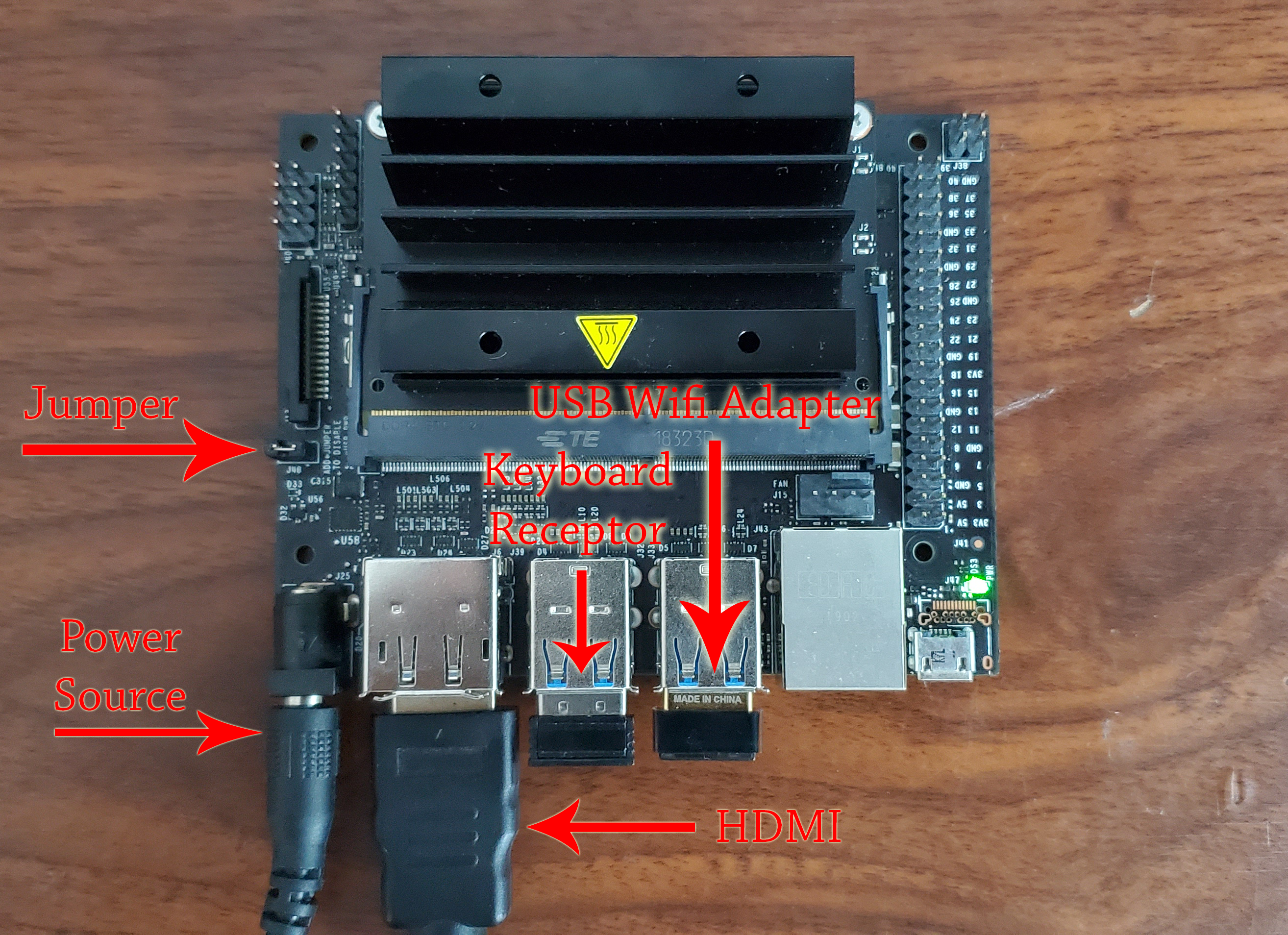
Because we power the Jetson Nano through an external 5-volt source with a Jack connector, we will have to place a Jumper on the J48 connector of the Jetson as shown in the image.
This is the explanation on how to install the Jetson SDK OS image on an SD card. You will need a computer with an SD card reader to install the image.
We recommend downloading the latest version of the SDK, in this guide I use version 4.3.0 (most recent to date).
Official Link: https://developer.nvidia.com/embedded/jetpack
You'll need to unzip the file to get the image file (.img) to write to your SD card. If you do not have a program to unzip, I recommend any of the following according to your operating system (windows in my case).
7-Zip (Windows):
The Unarchiver (Mac):
Unzip (Linux):
https://linuxize.com/post/how-to-unzip-files-in-linux/
Windows and Mac:
https://www.sdcard.org/downloads/formatter/index.html
Linux:
https://www.pcworld.com/article/3176712/how-to-format-an-sd-card-in-linux.html
My computer does not have an SD card reader so I use this external one (any reader is ok).
And this is the software for the SD card formatter. I especially like this program because this type of operating system creates multiple partitions in the SD memory and the format of them can be complicated if we want to reformat later, however this program does everything automatically.
You will need to use an image writing tool to install the image you have downloaded on your SD card. I recommend balenaEtcher as it works on all OSs and it is not necessary to unzip the.zip to perform the OS flash.
Download Link: https://www.balena.io/etcher/
Once the process is completed correctly, we see the following message.
Insert the SD into the SD slot of the Jetson Nano.
Connect the Jetson Nano to the screen using the HDMI cable, connect the wireless keyboard receiver, connect the network card and connect the power supply.
We will configure the operating system, it is very simple.
- Accept the terms.
- Select your language.
- Select your keyboard layout.
- Configure your wifi.
- Select your region.
- Select your credentials.
SUPER IMPORTANT NOTE: CHECK THE OPTION "Log in Automatically"
- Click ok to expand your Partition Size.
- Wait a couple of minutes.
- If everything works, you will see a screen like this.
- This video shows the final setup of the operating system.
- With this you should already have everything configured, from now on the HDMI cable and the wireless keyboard are no longer necessary. All programming and final setup will be done through SSH.
For this step we will create an ssh connection with the Jetson, if you have Mac or Linux are already preconfigured with OpenSSH library, so you can start your connection from the terminal with the following command.
ssh -L 8000:localhost:8888 youruser@youripIn my case the command is:
ssh -L 8000:localhost:8888 vic@192.168.0.28NOTE: it is also possible to activate this library in windows but I recommend using the instructions that I will show you next.
If you are a Windows user I recommend using the following program:
This animation shows how to configure Putty exactly as the last command.
Taking the Putty console as example, clicking on connect will display the following message.
Click "Yes" to bring up the following window, as long as you do not format the Jetson OS it will not appear again, at this time it will ask for the password that we defined in the previous section.
After inputting the password in the command console, this window will appear, indicating that we are already connected to the Jetson Nano.
Once the wireless connection to the console is established, we will have to copy and paste the following commands into it and execute them.
Command to download the project and get all the necessary files for the project.
git clone https://github.com/altaga/Anaphylactic-Skin-Reaction-Detection-during-ChemotherapyCommand to enter the downloaded folder.
cd Anaphylactic-Skin-Reaction-Detection-during-Chemotherapy/InstallerThis command will install all the libraries and configurations necessary to setup the project correctly. To facilitate its installation I'll make an.sh file that performs all this automatically, however I also attached the commands separately in Appendix A. Also the file can be reviewed by any text editor such as Notepad, Atom, VSCode, etc...
NOTE: Go for a coffee, some cookies and see the next chapter of your favorite series, because this process can take 45 minutes to 2 hours to complete, depending on your internet connection.
sudo bash Install.shWith this process we will have all the libraries installed correctly:
- TensorFlow 2.0
- Awscli (we haven't finished setup this library)
- Numpy
- Jupyter Notebook
- PahoMQTT
- OpenCV (No Contrib Version)
Once this process has concluded, we have to check that the Jupyter notebook works correctly, as it will be our UI for the rest of the tutorial. Next, write the following command:
jupyter notebookYou should see something like this in the terminal:
Copy the token that appears and without closing the window go to a browser and on the address bar input:
localhost:8000You should get a window such as this one:
Paste the token you copied previously:
If the token was valid we should have the Jetson files open on the browse, this is important because this window will allow us to manage the files easily, and allow us to execute the project's files.
AWS Jetson setup:First we have ti access our AWS console y look for the IoT core service:
Obtain your AWS endpoint, save it because we will use it to setup the JEtson and the webpage.
In the lateral panel select the "Onboard" option and then "Get started".
Select "Get started".
In "Choose a platform" select "Linux/OSX", in AWS IoT DEvice SDK select "Python" and then click "Next".
In Name write any name you'd like and then click on "Next step".
In the section, "Download connection kit for" press the button "Linux/OSX" to download the credential package (which we will use later) and click on "Next Step".
Click "Done".
Click "Done".
In the lateral bar, inside the Manage/Things section we can see our thing already created. Now we have to set up the policy of that thing for it to work without restrictions in AWS.
In the lateral bar, in the Secure/Policies section we can see our thing-policy, click on it to modify it:
Click on "Edit policy document".
Copy-paste the following text in the document and save it.
{
"Version": "2012-10-17",
"Statement": [
{
"Effect": "Allow",
"Action": "iot:*",
"Resource": "*"
}
]
}Once this is done, we will go to our pc and to the folder with the credentials previously downloaded and extract them.
We enter the extracted folder and we will rename the files the following way:
ThingNAME.cert.pem -> ThingCert.cert.pem
ThingNAME.private.key -> PrivateCert.private.keyNow, with the files already renamed we will go to our Jupyter Notebook in the following route:
In the right corner there's a button that says "upload"
By clicking on it we are able to upload our two certificates to the folder.
Click every single one of the blue colored "upload" buttons to finish the file upload.
By this point we should have all the necessary credentials.
AWSCLI Setup:This is the AWS library to manage and execute actions via Python for Cloud, so we have to set it up like so:
At the console we go to the IAM service.
In the Access Management/Users section we click on Add user.
We type any username and we click on "Next:Permissions"
Click "Attach existing policies directly", at the searchbar we write "S3" and we select the "AmazonS3FullAccess" policy.
We click on "Next" until we reach the success screen, where we will see the Access Key ID and the Secret Access Key, both keys we have to save in order to set up the Awscli.
From our Jupyter notebook UI at the "new" button open a new terminal.
Typoe the following command on it.
aws configureConfigure the credentials the following way:
AWS Access Key ID [None]: YOUR ACCESS KEY ID
AWS Secret Access Key [None]: YOUR SECRET ACCESS KEY
Default region name [None]: us-east-1
Default output format [None]: jsonReady! we have now configured the Jetson Nano.
Webpage Setup:Aws Credentials Setup:Enter the AWS console and search for the "Cognito" service.
Enter "Manage Identity Pools"
Enter "Manage Identity Pools"
Enter "Create new identity pool"
Type any name at the pool and check "Enable access to unauthenticated identities" and click on "Create Pool"
Just click "Allow".
We just got our POOLID, save it as we will use it afterwards.
Go to the AWS console and enter "IAM".
Inside the console enter the Role section, at the searchbar write "web" and enter in the one that says "Cognito_WebPagePoolUnauth_Role".
Inside the Role we click on the Attach policies button to add the services we need for our webapp.
Inside that window we need to add three services:
- AmazonS3FullAccess
- AWSIoTFullAccess
- AmazonDynamoDBFullAccess
Now that we have the permissions required we part ways to configure the database where we will have the patient's information. From the AWS we will search for DynamoDB.
Select Create Table.
Create a table with the following parameters, it is important that the names are the same ones we are showing in the image.
Name: HacksterDB Partition Key: PartKey Sort Key: SortKey
Once the table is created we can generate registers on it about patients which we will be able to visualize on our platform, the registers have to follow the following structure.
{
"Age": "56",
"App": " 03/03/2020",
"Cancer": "Sarcoma",
"Comments": "Entrepreneur, if you don't have at least one TitanRTX on your computer, don't talk with him",
"Incidents": "1",
"Medicine": "Carboplatin",
"PartKey": "dev1",
"SortKey": "Jen-Hsun Huang"
}Description of the registers:
- Age: age of the person
- App: Date of his following appointment
- Cancer: Type of Cancer
- Comments: Any comments of the specialist
- Incidents: Number of incidents to date.
- Medicine: Pharmacological treatments
- PartKey: The device that procures the register
- SortKey: Name of the patient
Finally we will create an S3 bucket which will allow us to store any file or image we need. From the AWS console look for the S3 service.
On S3 click the button to create a bucket.
Type any name for the bucket but remember it as we will call it back afterwards.
Uncheck all the block options as in the image:
Once all that is finished, we have everything ready to setup our webapp.
Uncheck all the options to block as in the image:.
With this done we have created our bucket, with the following URL.
https://yourbucketname.s3.amazonaws.com/Download the Github file to your PC.
Inside the project folder go to: ReactAPP\src\views\examples.
With your favourite editor open the following files:
- aws-configuration.js
- MyCard.jsx
- Card.jsx
- Profile.jsx
Inside "aws-configuration.js" paste our POOLID and our AWS Endpoint.
Inside "MyCard.jsx" paste your bucket URL.
Inside "Card.jsx" paste your bucket URL.
Inside "Profile.jsx" paste the name of the DB, if you named it "HacksterDB" you don't need to do anything else.
To visualize the DB in a Navigator you need to install NodeJS in your computer.
Link: https://nodejs.org/es/
Once installed enter the folder of the project called "ReactAPP".
Once there, oper the terminal or in the case of windows cmd.
NOTE: If you are using windows just type cmd on the search bar.
In the cmd or terminal write the next command.
npm installAfter all the dependencies have been installed, at the console write:
npm startEnter the Jupyter notebook UI from the browser at "localhost:8000". The token should not longer be needed.
Enter the "Anaphylactic-Skin-Reaction-Detection-during-Chemotherapy\Jupyter Notebook\Anaphylactic Skin Reaction Detection during Chemotherapy.ipynb" folder
With everything set up we now enter the browser before performing the code revision we need to paste our Bucket name and AWS IoT Endpoint.
Product:
Essential components:
UI:
- Real time model performance
- Real time emergency notifications
- Patient database search tool
In summary in this project we have:
- Developed our own Machine Learning algorithms and procedures for a particular problem, implementing Computer Vision on Tensorflow.
- Used the Nvidia Jetson Nano to its full capabilities.
- Fully documented the whole process and made sure that all the present documentation can run at any time in any Jetson nano with a setup such as this one.
But, the most important part is that it solves a real problem. When I started the project I didn't want to just do a weekend project or just a very cool robot. What I wanted is to work backwards from the problem, then looking at what hardware I needed to solve it. Thankfully because of my education as a Biomedical Engineer and the work I do in conjunction with Hospitals, clinics and healthcare institutions I have a good grasp of certain problems in some areas. This particular one (the allergic reaction to chemotherapy and a prompt reaction) was indeed one that was asked of me to develop a proper solution to. And the fact that it fit with one of the Un's SDGs (Sustainable development goals) made it much better.
And I actually think that the project (for a prototype) is almost ready for testing and even going beyond with it. Improvements I could mention are only on the aesthetic side (I'm thinking an IP camera-type enclosure). This also meaning to develop a much more commercial product.
Thank you for reading and keep hacking!
References:Links:
(2)https://www.ncbi.nlm.nih.gov/books/NBK333506/table/ch04.sec1.table1/
(3) https://medium.com/@alexrachnog/using-caffe-with-your-own-dataset-b0ade5d71233
(4) https://www.pyimagesearch.com/2017/08/21/deep-learning-with-opencv/
Appendix A:Install.sh Content:
export PATH=$PATH:~/.local/bin
sudo apt-get update
sudo apt-get upgrade -y
sudo apt-get install libhdf5-serial-dev hdf5-tools libhdf5-dev zlib1g-dev zip libjpeg8-dev -y
sudo apt-get install python3-pip -y
sudo pip3 install -U pip testresources setuptools
sudo pip3 install -U numpy==1.16.1 future==0.17.1 mock==3.0.5 h5py==2.9.0 keras_preprocessing==1.0.5 keras_applications==1.0.8 gast==0.2.2 enum34 futures protobuf
sudo pip3 install --pre --extra-index-url https://developer.download.nvidia.com/compute/redist/jp/v43 tensorflow-gpu
sudo pip3 install notebook awscli paho-mqtt
sudo apt-get install python3-matplotlib python3-opencv python3-scipy -y














Comments
Please log in or sign up to comment.