See what's new at Remyx.ai
IntroductionRoughly 1% of the population lives with anxiety around their next seizure event. Anti-convulsant medication does not help at least 20% of those afflicted, but also carries negative side effects in high doses. This fear stands in the way of performing everyday tasks like driving a car or swimming. There exist devices that can detect that a seizure is occurring, but we aim to build a wearable EEG device that can predict that a seizure is about to occur so a caregiver and/or the user can pause what they are doing and take appropriate preventative action.
Five years ago, a Kaggle contest framed the challenge of developing a machine learning algorithm to help predict seizure events. Contestants were provided 106 GB of training data in.mat files. Each training sample represents 10 minutes of raw intracranial EEG readings. For 5 dogs, data included 16 channel readings sampled at 400 Hz. For 2 human patients, data was sampled at 5000 Hz over 24 channels.
Here is a short segment sampled from one channel:
Medical research supports the concept of 4 distinct phases corresponding to: between, preceding, during, and after seizure events. For the purposes of this prediction task, the main challenge is to develop a binary classifier to differentiate the so-called interictal (between events) and the preictal (precede events) states.
Most contestants engineered lab-tested features based on FFT or time-correlation between the channels with the results of a similar contemporaneous contest in mind.
Unconstrained by the real-world limitations of computational feasibility on embedded hardware, the winning models involved complex ensembles trained with various machine learning algorithms. The contest finished in 2014, nearly a year before frameworks like Tensorflow were available to enable deep learning experiments.
Since the contest, applying convolutional neural networks (CNNs) to spectrograms has been a popular approach to speech and signal processing tasks. This borrows from the successes in computer vision by reframing the problem to identify visual signatures called formants in the time-frequency representation of a signal.
In addition to the benefit of regularization achieved through Fourier Transform, the FFT algorithm is very fast and can be performed on resource-limited hardware.
CNNs too, have been getting smaller and so we combine these two powerful algorithms to run inference on embedded hardware in real-time to predict seizures through identifying preictal states.
Training Our Neural NetWe begin with a few simplifying assumptions.
Due to resource limitations, we do not want to read 10 minutes of 16/24 channel EEG recordings into memory. Instead, we will break a sample into 200 3-second segments. Implicitly, we assume that each is equally well representative of the signatures we expect to learn in distinguishing whether the segment precedes a seizure event or not.
Further, we assume each channel is independently representative of these signatures. This allows us to expand the dataset at the cost of disregarding the covariance between channels. First, we tried to visualize the spectrogram under the standard FFT:
By experimenting with different parameter choices and variants of FFT, we settle upon the Short-Time Fourier Transform (STFT).
To avoid recomputing these transforms, we output an image to file. Here is an example after applying additional transformations and normalization to surface visually discernible patterns in the frequency representation. We resize the image to 128x128 to load more images and train faster.
To learn more about our data processing, see our blog post.
To quickly explore the idea that CNNs will bring something useful, we set a baseline using very simple NN architecture with the Keras sequential API. Our vanilla CNN includes 5x5 convolutional filters, followed by a 3x3 conv layer before flattening and funneling through fully connected layers with ReLU activations, and dropout for additional regularization.
def build_network():
model = Sequential()
model.add(Conv2D(32, (5, 5), input_shape=(128, 128, 1)))
model.add(Activation('relu'))
model.add(MaxPooling2D(pool_size=(2, 2), strides=(2, 2)))
model.add(Conv2D(32, (3, 3)))
model.add(Activation('relu'))
model.add(MaxPooling2D(pool_size=(2, 2), strides=(2, 2)))
model.add(Conv2D(32, (3, 3)))
model.add(Activation('relu'))
model.add(MaxPooling2D(pool_size=(2, 2), strides=(2, 2)))
model.add(Flatten())
model.add(Dense(2048))
model.add(Activation('relu'))
model.add(Dropout(.5))
model.add(Dense(512))
model.add(Activation('relu'))
model.add(Dropout(.5))
model.add(Dense(256))
model.add(Activation('relu'))
model.add(Dropout(.5))
model.add(Dense(1))
model.add(Activation('sigmoid'))
adam = Adam(lr=0.0001, beta_1=0.9, beta_2=0.999, epsilon=1e-8)
model.compile(optimizer=adam, loss='binary_crossentropy', metrics=['accuracy'])
return modelDue to extreme class imbalance, we upsample the preictal examples.
Calling the fit method, we see signs of learning in the training output using a couple thousand samples. To scale up training, we switch to the fit_generator method to iterate through our collection of nearly 10 million images.
We find our model is underfitting, as evidenced by the plateau in loss with similar training and validation performance:
Epoch 4/100
128/128 [==============================] - 525s 4s/step - loss: 0.6798 - acc: 0.5515 - val_loss: 0.6768 - val_acc: 0.5682
Epoch 5/100
128/128 [==============================] - 509s 4s/step - loss: 0.6769 - acc: 0.5668 - val_loss: 0.6775 - val_acc: 0.5579
Epoch 6/100
128/128 [==============================] - 532s 4s/step - loss: 0.6761 - acc: 0.5624 - val_loss: 0.6732 - val_acc: 0.5728
Epoch 7/100
128/128 [==============================] - 504s 4s/step - loss: 0.6748 - acc: 0.5714 - val_loss: 0.6706 - val_acc: 0.5803
Epoch 8/100
128/128 [==============================] - 505s 4s/step - loss: 0.6737 - acc: 0.5686 - val_loss: 0.6696 - val_acc: 0.5791
Epoch 9/100
128/128 [==============================] - 504s 4s/step - loss: 0.6733 - acc: 0.5665 - val_loss: 0.6685 - val_acc: 0.5808
Epoch 10/100
128/128 [==============================] - 504s 4s/step - loss: 0.6674 - acc: 0.5834 - val_loss: 0.6670 - val_acc: 0.5852
Epoch 11/100
128/128 [==============================] - 504s 4s/step - loss: 0.6676 - acc: 0.5782 - val_loss: 0.6669 - val_acc: 0.5863
Epoch 12/100
128/128 [==============================] - 503s 4s/step - loss: 0.6667 - acc: 0.5803 - val_loss: 0.6655 - val_acc: 0.5884
....
Epoch 99/100
128/128 [==============================] - 502s 4s/step - loss: 0.6242 - acc: 0.6316 - val_loss: 0.6317 - val_acc:
0.6271
Epoch 100/100
128/128 [==============================] - 521s 4s/step - loss: 0.6253 - acc: 0.6318 - val_loss: 0.6266 - val_acc:
0.6321Nonetheless, we can begin to pin down some of the details around our simplifying assumptions and explore more complex models.
Next, we are trying transfer learning by training a model based on the inception_v3 architecture which was pretrained on ImageNet, using TF-Slim.
The inception_v3 architecture has much higher capacity to learn patterns in the data than our simple architecture above. With fine-tuning, many image features will be suitable for our task even though the image set is quite different.
After training for some time, our model reaches 72% validation accuracy on a balanced subset of nearly 1 million spectrogram images. We're well-convinced to invest the time & effort into training and evaluating a model like this on more data, once we have it running fast on the pi.
Also, we might try unsupervised pretraining since half of the available Kaggle data consists of testing samples.
Update: Using newer apis, different model architectures, and slightly different preprocessing we reach much higher validation accuracy in this jupyter notebook.
Hacking EEG Devices
We tried the Muse headset since an Adafruit tutorial described it as a straight forward hack. Unfortunately, the manufacturers were unable to continue supporting this and the referenced library appears to be defunct.
We also found the more affordable Force Trainer. However, the firmware performs FFT and returns values within different bands in the power spectrum. Instead, we need the raw EEG signals like those we used to develop a machine learning model to analyze.
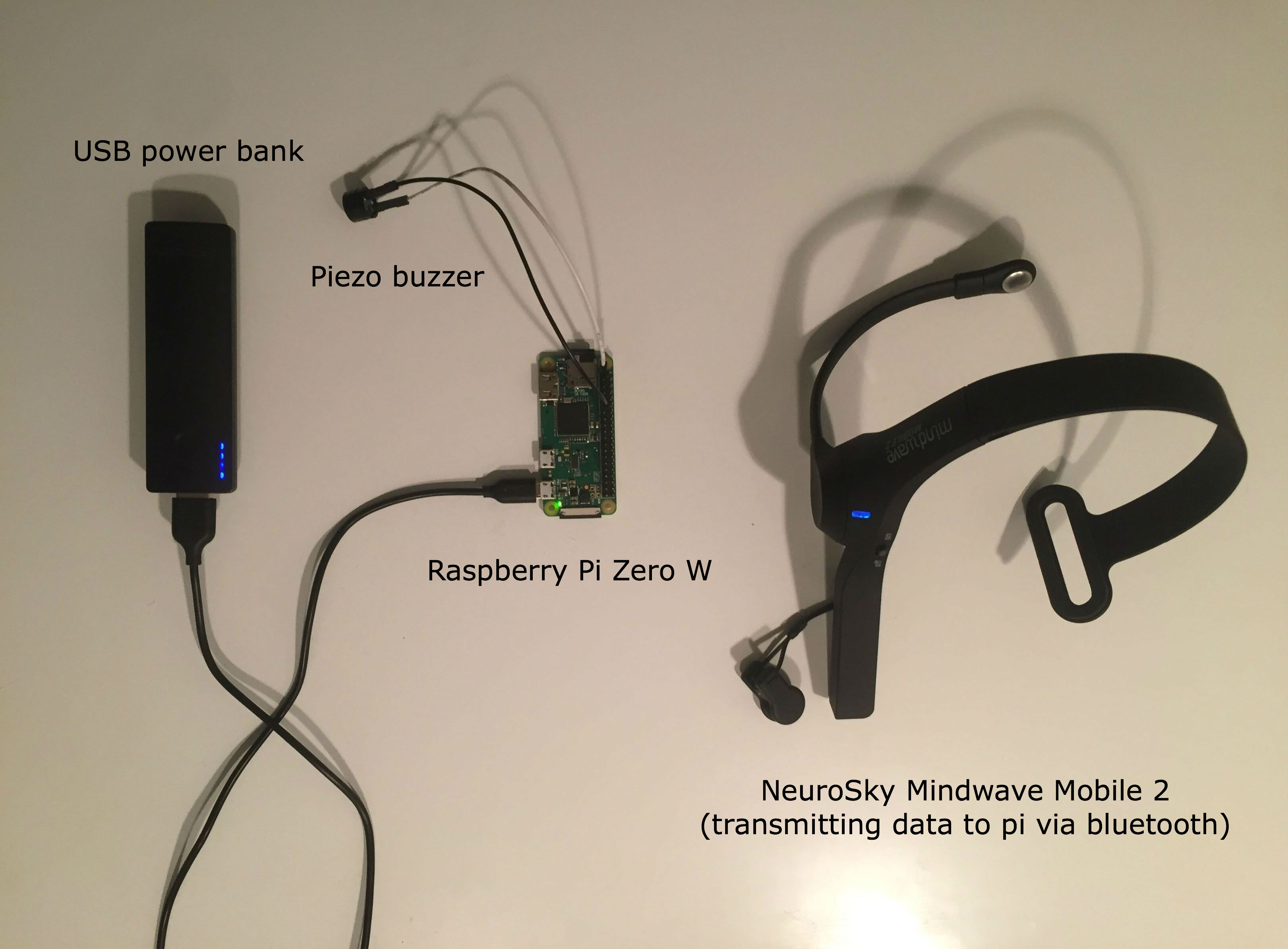
Finally, we've settled on the NeuroSky MindWave Mobile device. With a price between the two options above, while supporting the collection of raw EEG signals, this product appears promising. We used this repo to read the raw EEG values from the headset.
import bluetooth
from mindwavemobile.MindwaveDataPoints import RawDataPoint
from mindwavemobile.MindwaveDataPointReader import MindwaveDataPointReader
import textwrap
if __name__ == '__main__':
mindwaveDataPointReader = MindwaveDataPointReader()
mindwaveDataPointReader.start()
try:
if (mindwaveDataPointReader.isConnected()):
while True:
dataPoint = mindwaveDataPointReader.readNextDataPoint()
if (dataPoint.__class__ is RawDataPoint): #Only want the raw vals
print(dataPoint) #printing as a test
else:
print((textwrap.dedent("""\
Exiting because the program could not connect
to the Mindwave Mobile device.""").replace("\n", " ")))
except KeyboardInterrupt:
sys.exit()This headset will stream the data to our raspberry pi via bluetooth. Then the pi will format the data like our training samples and run inference on it to predict the onset of a seizure.
Running Inference
We apply our signal processing pipeline on device before rendering an image.
Finally, we run inference on the spectrograms to raise an alert (via a piezo buzzer in our prototype) to the user if a signature corresponding to an impending seizure event has been detected.
To reduce false positives, we need to develop a post processing workflow so that users are not exhausted by model errors. One simple idea would be to raise an alert only after the count of positives over some time window exceeds a empirically determined threshold.
With a device, we can perform data collection that will help to refine our models. To further reduce costs, consider building an EEG circuit, perhaps with fewer electrodes.
In the future, we plan to put the model on a smaller low power device like a Blue Pill, which can run on a coin cell battery.
Final ThoughtsWith easy access to resources like cheap hardware, as well as open-sourced machine learning algorithms and data, it is an exciting time to participate in the conceptualization and development of medical device technologies.
We hope to inspire others to productize the life-changing technologies of tomorrow.
Check out our repo on this project here!
















Comments