This project will help guide you in creating an artificial intelligence-powered device to allow you to directly interface your brain with your own hardware and software projects. Yep, a real AI-Powered Brain-Computer Interface!
Along the way, you will learn a bit about brain electrophysiology; neural oscillations; signal processing, interfacing with low-level USB devices; threading with Python; building Neural Networks with PyTorch; making awesome GUI's in Python with OpenGL using Pyglet, and of course awesome techniques in unsupervised learning, clustering and anomaly detection using artificial intelligence!
We're not just going to re-use an off-the-shelf computer vision architecture either (not that there's anything wrong with that), we'll be building a neural net from scratch, so you can learn how they work from the ground up.
Why?Deep learning is revolutionizing many fields of science and technology. By using machine learning, we can predict and classify natural processes without building explicit models - something vital when the data is hard to interpret!
The most complex object in the known universe is something we all carry around with us every day - no, not our smartphones - it's our brains. But understanding what going on in there is hard, as our subjective reflections can't describe what's happening inside our own skulls.
There are non-intrusive technologies we can use (because who really want to drill holes in their skulls before Elon Musk's Neuralink is complete), such as MRI, EEG, and FNIR. But of these, only EEG is accessible to us hacksters, via products such as the NeuroSky MindWave or Star Wars Force Trainer. But the processing power of such devices is so low, that they have to be hooked up to a PC or can only extract single brain wave frequencies, and ignore the vast majority of complex signals being generated by the brain.
With the power of the Jetson Nano, we can finally do deep and complex analysis of EEG data, directly on a portable device connected to the internet! This project will bring us to the very cutting-edge of what currently possible in EEG analysis.
How?Electrophysiology
Brains do operate with electrical signals, but not at all in the same way our electronic devices operate. There's a heck of a lot more going on in the brain than just electrical signalling, but let's stay focused on that aspect. The human brain contains 86 billion neurons, with each of those neurons receiving input from multiple other neurons in a network.
Each neuron acts like a signal-processor on these inputs; when a neuron has decided that it has received enough inputs within a certain time-frame, it 'fires' an electrical pulse to neurons further down the network. This is an all-or-nothing response - downstream neurons only know that a neuron has fired, they don't get any other information. The action potential is pretty fast too; the peak voltage pulse is over in about a millisecond.
The brain doesn't have a centralized 'core' like the CPU of a computer. The neurons of the brain operate in parallel, giving it huge compute and bandwidth. But each neuron needs to do signal-processing on multiple inputs simultaneously, so it would be better to have the inputs be coordinated. The brain orchestrates this by operating in cycles, with neurons all firing in bursts to maintain synchronization (if they need to fire at all).
We can't detect these fast and teeny-tiny action potentials though skull, flesh, and skin, but in aggregate, we can measure the local field potential reflecting their summed activity. The fields are of course damped and smeared by all the meat and bone between our sensor electrodes and the brain, so it's hard to get a localized or even very accurate signal, but amazingly, we can still measure our brain activity!
During alert states, the brain operates at higher frequencies than when relaxed or various sleep phases. You can't directly detect these internal states consciously, but by recording and processing them, we have a 'biofeedback' loop that lets you actively learn to control your brain state!
Electroencephalography
The device needed to record the brain's electrical activity is actually pretty simple. The electrical activity of animal brains was first recorded nearly 150 years ago! Nowadays, the recording is obtained by placing electrodes on the scalp, and the signal is amplified by customs ICs. It should be possible to build a nice EEG device based around a Texas Instrument ADS1299 and read directly via SPI from the Jetson Nano, but for this project, I don't have enough time for circuit development when there's Artificial Intelligence engineering to do!
For this project, I will be using an old OCZ Neural Impulse Actuator (NIA). The internals has been nicely documented here. This device can be bought cheaply on certain auction sites on the internet, and it acts as a USB HID device; data can be easily read as a stream of serial data. Newer EEG devices will be able to use the same awesome Jetson Nano platform with the code presented here - you just need a way to receive streaming EEG data. The biggest benefit with newer devices is in the number of concurrent channels the EEG device provides.
Setting up the Hardware and Software
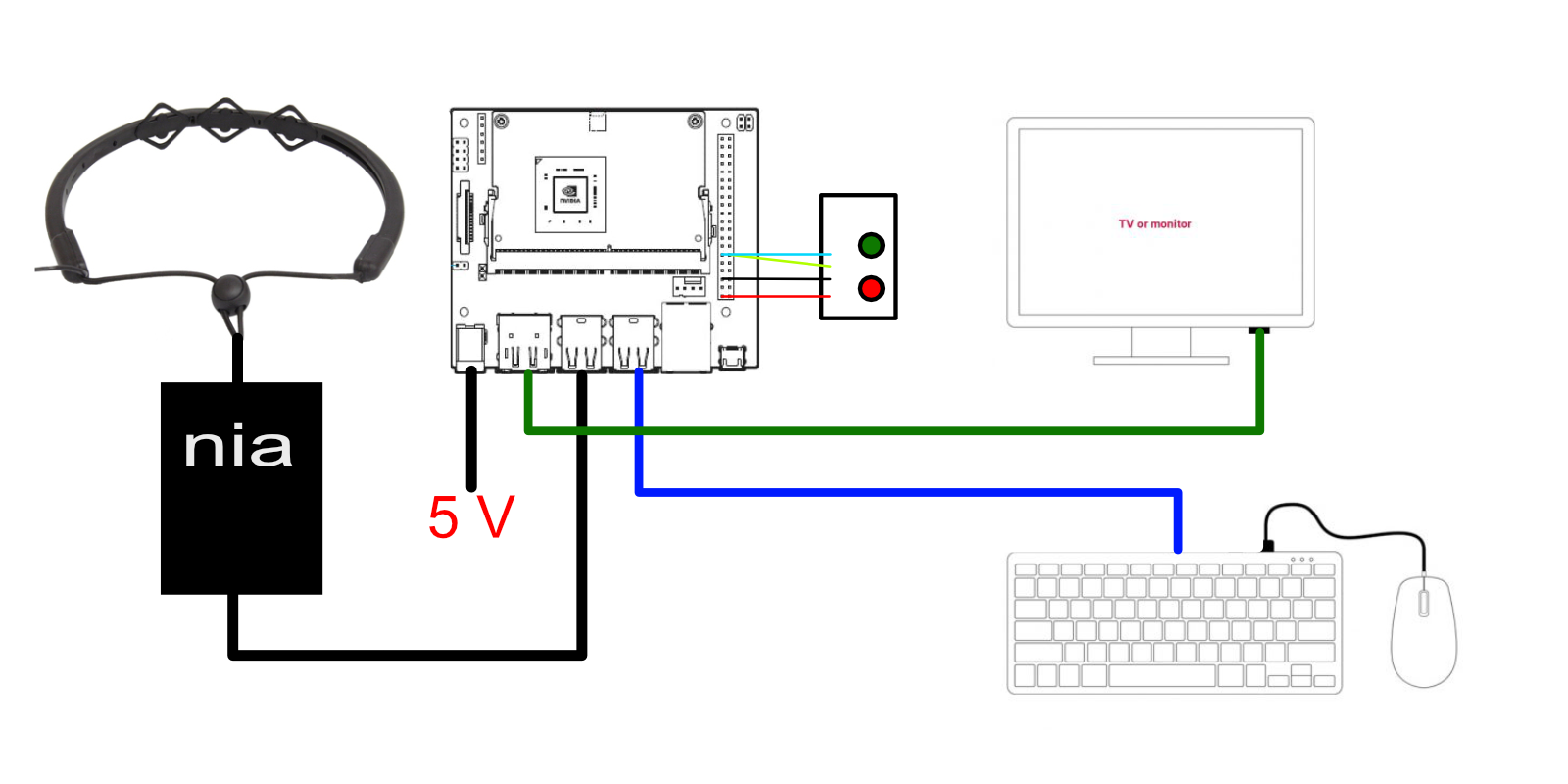
Unpack your shiny new Jetson Nano, and use the barrel jack for power (the power draw on the NIA is pretty low, but we want to do a lot of processing). You can find the setup information here. Make sure you have a big enough SD card on hand! (I forgot to order one, and had to sacrifice a Raspberry Pi formatted sd card)
The OCZ NIA can be plugged into the Jetson's USB sockets, along with a mouse and keyboard. Make sure you have a compatible WiFi adaptor or Ethernet cable handy to get yourself online, as we need to download lots of packages!
First, we have to set up our development environment, and as it's 2020, so let's do this in Python 3 with the latest updates!
Let's update apt to get things started:
sudo apt updateThen we can install and upgrade Python 3 and libraries we'll need later:
sudo apt upgrade python3 libusb-dev
sudo apt install -y git cmake build-essential libopenblas-base libatlas-base-dev gfortran python3-dev libhdf5-serial-dev hdf5-toolsAs we will be using just Python 3 and its version of Pip from now on (and so should you), let's make it the default.
cd ~
wget https://bootstrap.pypa.io/get-pip.py
sudo python3 get-pip.py
rm get-pip.py
echo "export PATH=$PATH:/$HOME/.local/bin" >> .bashrc
echo "alias python=python3" >> .bashrc
echo "alias pip=pip3" >> .bashrc
source .bashrcNext, we can install a bunch of Python packages to make and present the analysis. This step is going to take quite a while, around an hour or so. I like verbose mode, so I can see all the work the Jetson is doing ;)
pip install --user --verbose pybind11 Cython jupyter numpy matplotlib pyglet pyusb scipy sklearnFinally, for the Autoencoder, we'll need Pytorch, then we are done!
wget https://nvidia.box.com/shared/static/ncgzus5o23uck9i5oth2n8n06k340l6k.whl -O torch-1.4.0-cp36-cp36m-linux_aarch64.whl
pip install --user torch-1.4.0-cp36-cp36m-linux_aarch64.whl
rm torch-1.4.0-cp36-cp36m-linux_aarch64.whlStick the electrode-band on your forehead, and it's time to move on...
Signal Processing
If you want to process EEG data, typically a Fourier Transform is used to analyze the signal, and determine which state the brain is in. It is a fast way to determine what mix of frequencies are present in a recording of the brain's EEG signal.
But, of course, it's not so easy... the EEG information is mixed up with lots of other signals, and most of them are much stronger than the tiny brain-derived signals we want to extract - muscles produce slow but massive signals and mains voltages produces a constant background signal of 50 or 60 Hertz, depending on where you live.
Using the software installed above, you can now use the Jupyter notebook in the project GitHub to look at the raw data! As the USB library is acting a bit weird, it needs root access. Start the Jupyter server with:
sudo jupyter notebook --no-browserand then open up the.ipynb file from the GitHub repository.
You can just the first few cells, which loads up the needed packages, and defines our EEG class.
We are using PyUSB to access the OCZ NIA. It streams data that looks like this (and is described here):
array('B', [254, 68, 128, 68, 59, 128, 42, 44, 128, 209, 13, 128, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 56, 189, 166, 46, 244, 143, 4])
array('B', [96, 234, 128, 25, 213, 128, 10, 161, 128, 197, 173, 128, 72, 186, 128, 130, 240, 128, 178, 26, 129, 9, 28, 129, 161, 24, 129, 48, 210, 128, 9, 182, 128, 187, 116, 128, 152, 66, 128, 190, 67, 128, 12, 27, 128, 107, 229, 127, 56, 189, 136, 214, 230, 55, 16])
array('B', [29, 83, 128, 162, 105, 128, 111, 120, 128, 219, 138, 128, 175, 142, 128, 122, 160, 128, 157, 205, 128, 239, 212, 128, 232, 10, 129, 224, 8, 129, 200, 58, 129, 46, 70, 129, 132, 23, 129, 0, 18, 122, 0, 18, 122, 0, 18, 122, 56, 189, 198, 15, 49, 113, 13])
array('B', [243, 6, 129, 163, 238, 128, 219, 242, 128, 51, 226, 128, 180, 223, 128, 149, 244, 128, 54, 213, 128, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 56, 189, 198, 15, 56, 113, 7])The protocol is described in the link, and the important things are:
- the last position is a count of the valid EEG samples
- the data is stored in 3-bytes, least significant byte first.
- empty samples are [0, 18, 122], and can be ignored
i.e. for this packet:
array('B', [254, 68, 128, 68, 59, 128, 42, 44, 128, 209, 13, 128, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 0, 18, 122, 56, 189, 166, 46, 244, 143, 4])There are 4 EEG samples, [254, 68, 128], [68, 59, 128], [42, 44, 128], and [209, 13, 128].
We can decode the bytes in Python 3 with the 'from_bytes' command.
>>> int.from_bytes(bytearray([254, 68, 128]), byteorder='little')
8406270With all that sorted, it's time to take a look at a raw signal taken off the NIA!
This signal shows huge and slow oscillations cause by eye movement, overlaid with a constant 50 Hz hum from the power lines. What we actually want is the pure brain wave signals, which range from about 2-40 Hz. We can remove the unwanted signals with a band-pass filter. As the name suggests, it will only allow a 'band' of the frequencies through, and the unwanted signals above and below the cutoffs will be blocked. We can visualize the low and high frequencies we want to remove too:
After passing the same data through the bandpass filter, we end up with this kind of data, a nice clean signal for further processing!
This cleaned-up signal captures the signals produced by the brain from 5 to 35 Hertz. A simple way to analyze the data if with a Fourier Transform, which breaks the signal down to the strength of the frequencies in the sample. Doing it with our data gives us this kind of output:
We can see the signals above 35 and below 5 in our bandpass-filtered data are mostly gone now. The bleed-through can be modified by changing the steepness of the bandpass filter, the 'order' of the Butter-bandpass filter.
You can use the Jupyter notebook to capture some brain wave data. Try with while relaxing with your eyes closed, and again, eye's open thinking about all the things you have learned.
The last thing the Jupyter does is allow us to capture data for training later. It's set up to capture data in 5-minute blocks, and we need about a 1 Mb per second to store our raw EEG data.
The Fourier transform is a nice first-start in analyzing brain activity, but we know signals blend into one another, vary in intensity and form complex secondary patterns over time. This type of signal processing loses timing signals. Something like Wavelet analysis might help, but we would still need to further classify the output of the wavelet analysis. Just getting a list of frequencies and positions can't possibly capture what state our brains are in. We need an analysis technique with more power than a simple Fourier Transform. That's where we can use cool machine learning techniques, AKA:
Artificial IntelligenceOne way to process the complex time-series data streaming from our EEG device is to cluster the data into categories. Not just the dominant brain frequency (which would vary from person to person), but a 'fingerprint' of the type of brainwaves produced during different mental activities.
A second's worth of data (4096 data points or 'dimensions') is a lot to try and analyze, so we should reduce this into something both deeper and more simple. The Fourier Transform does this by reducing the signal into its component frequencies, but what we really want to do is to classify the 'brain-states' that generated the signals in the first place!
First, we can reduce the dimensionality of the problem with an Autoencoder. This kind of neural network performs a seemingly silly task - generating an output that looks justlike the input!
The 'trick' is that the data is bigger than an internal bottleneck within the neural network. The only way the neural network can regenerate the input data is if it can 'compress' the data to fit through the bottleneck. This compression reduces the number of dimensions of the data, and as a bonus filters out noise (as there is no way a neural network can predict noise, this architecture is sometimes referred to as a 'denoising autoencoder').
Once our data is compressed into a lower-dimensional representation, we can both visualize the EEG data, and perform clustering to identify common states! One such clustering method is the k-nearest neighbors (k-NN) algorithm. This fast algorithm lets of quickly identify the brain state from the time-series EEG data and the GPIO's on the Jetson board allow us to use these states to interact with the physical world!
PyTorch
In the Github Repo is a Jupyter Notebook that generates an Autoencoder Neural Network. There are several things we need to do to train up a network:
- Prepare the data
- Divide the data into Training and Testing data
- Design the network, layer-by-layer
- Decide on how the what metric we evaluate the network
- Decide on how we optimize the model to fit the data
- Finally, decide on hyper-parameters
The Notebook goes over each of these points, but let's take a look at a few in detail!
Create the Autoencoder
Our Network should look like an hourglass: wide data comes in, goes through a few layers, where it gets compressed, and then the compressed layer is expanded until its the same size as the input.
In PyTorch, we first predefine the layers we want to use in the __init__ method. We can then use them how we want. In this case, I split the compression and expansion processes into two methods, 'encode' and 'decode'. This way, we can later just use the 'encode' half to process EEG data. Both methods are called in a 'forward' method that we use to run the entire process during training and testing.
class AE(nn.Module):
def __init__(self, ae_width):
super(AE, self).__init__()
self.fc1 = nn.Linear(1024, 256)
self.fc2 = nn.Linear(256, 64)
self.fc3 = nn.Linear(64, ae_width)
self.fc4 = nn.Linear(ae_width, 64)
self.fc5 = nn.Linear(64, 256)
self.fc6 = nn.Linear(256, 1024)
def encode(self, x):
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
return self.fc3(x)
def decode(self, x):
x = F.relu(self.fc4(x))
x = F.relu(self.fc5(x))
return self.fc6(x)
def forward(self, x):
x = self.encode(x.view(-1, 1024))
return self.decode(x)Loss Function
After we first pass a sample through our autoencoder, the results will be bad - actually random! That's because the network weights are set to be random at startup.
To train the network, we first need to be able to know how badly it's doing. The mean squared error (MSE) is the most commonly used loss function for regression. Mean squared error is calculated as the average of the squared differences between the predicted and actual values. The result is always positive regardless of the sign of the predicted and actual values and a perfect value is 0.0. The squaring means that larger mistakes result in more error than smaller mistakes, meaning that the model is punished for making larger mistakes.
In the Jupyter Notebook, this is defined in the 'train' function with:
loss = F.mse_loss(recon_batch, data.view(-1, 1024), reduction='sum')Luckily for us, PyTorch does most of the work, and 'mse_loss' returns the loss for us when we give it the input and output of the neural network data.
Optimizer
Once we know how wrong our Neural Network was, its time to figure out what to do about it. This article doesn't have time to go into any depth on back-propagation, except to say that's it's linear algebra and calculus, not magic that makes AI work.
Here's a cool picture to illustrate how the optimizer works though:
Basically, the job of the optimizer is to take a look at the results of the optimization so far and decide what to do next to get an even better result. A commonly used optimizer and one that's included in Pytorch is called Adam. We instantiate it in the Notebook, with a learning rate parameter. If we imagine Adam is telling us which direction to go in, the learning rate is how big the step should be.
optimizer = optim.Adam(model.parameters(), lr=1e-4)Learning
Then it's just a matter of cycling over the EEG data (each cycle through the entire dataset is called an epoch), and tuning the autoencoder weights until the output looks like the input! The Notebook generates a plot showing the input and output of a random EEG data sample every 5 epochs.
Let's take a look as see how well it did:
After 50 epochs, the Autoencoder has learned to reconstruct the data pretty well! And this is through a bottleneck, i.e. we have learned to compress an EEG trace of 1024 values to just 24 numbers! These 24 numbers represent a 'brain-state' we can now process for feedback or to use as an input to control software or hardware.
Clustering and Anomaly Detection
Autoencoders learn from the data they have seen during training. So, what happens when they are shown a very different kind of input than what they are used to? Well, they have no idea how to reconstruct that data, so the output will look very different from the input
Remember that MSE loss function we discussed earlier? Well, if we see a high MSE score on new data, we know its something the Autoencoder has never seen before and is just 'guessing' an output. That's a really powerful feature of Autoencoders: they learn what's 'normal' just from seeing a bunch of data.
It's often really hard to write a program to detect unusual patterns in data when each individual data point is in a normal range. For example, how would you detect an unusual sound in a forest? With an autoencoder, you just train it on recordings of forest sounds, and to detect weird noises, you compare the sound waveform inputs and outputs of the autoencoder using MSE!
So now our simple EEG device is an AI-powered medical device to detect unusual brain wave patterns! Studies have shown that before people have epileptic seizures, they having unusual brain wave patterns. But knowing the exact pattern that precedes a seizure is extremely hard to identify... so let's not do that, but instead, let's use our Autoencoder to warn patients as soon as anything unusual is detected!
Clustering is another way we can use an Autoencoder. We know that the brain produces different brain wave patterns in different states, and we also know we can now capture the brain wave state in a vector of twenty numbers. Using an algorithm like k-means, we use machine learning to automatically find these states.
Now that we have hooked up the EEG to the Jetson Nano to captured data, its time to tie it all together in a real-time application.
The first issue to deal with is in capturing and processing data. The processing takes time, tens to hundreds of milliseconds, but we have EEG data streaming in at 4096 samples a second, and we can't afford to lose any, or we risk corrupting our data.
So, we'll have to use Python threading to interleave data processing with data capture. Pythons Global Interpreter Lock prevents the interpreter from doing more than one thing at a time, even if there are multiple cores available. We could use Multiprocessing to use more cores, but even processing four thousand samples a second is easy for our processor, so there is more than enough time to do the required data processing in the gaps between capturing samples.
andThe way our application will work is to loop through these processes
- Start a data acquisition thread
- Signal process the previous data sample with the band-pass filter
- Pass the filtered data through a Fourier Transform
- Generate and display images of the data
- Complete the data acquisition
For this example, we will create a line graph of the EEG data, a Waterfall plot and a histogram of the brain waves, all on OpenGL on the Jetson Nano. ThePyglet library will handle the OpenGL work for us, it's a great library to use for graphics and sound, and even for making your own games!
You can run the finished app with:
sudo python3 app.pyThe code is on the AIoT Github repository linked with this project! Running it with an OCZ NIA will produce this kind of result in a new window:
On the left is a Waterfall plot of the Fourier transform data. On the bottom right is a filtered trace of the 250 milliseconds of EEG data. On the top right is another view of the Fourier transform data, in the form of a bar chart, low frequencies on the left, higher on the right between 5 and 35 Hertz. The height of the bras chart is the intensity.
If you don't have an NIA device yet, don't worry. If one is not detected, the app will still run using randomly generated data, so you can learn how everything works!
Let's go through the important bits of the app! First, the very two lines of the app:
andpyglet.clock.schedule(update)
pyglet.app.run()The first line tells Pyglet to run the 'update' function repeatedly, and the second line hand control of the program to Pyglet, which will then run until you close the window.
The 'update' function looks like this:
def update(x):
# 1
window.clear()
# 2
data_thread = threading.Thread(target=nia_data.get_data)
data_thread.start()
# 3
fourier_data,steps = nia_data.fourier()
# 4
waterfall = pyglet.image.ImageData(160,248,'I', fourier_data)
waterfall.blit(20,20)
# 5
for i in range(8): # this blits the brain-fingers blocks
for j in range(steps[i]):
step.blit(i*50+210, j*15+180)
# 6
graph = nia_data.waveform()
graph_image = pyglet.image.ImageData(410,140,'RGB', graph)
graph_image.blit(210,20)
# 7
data_thread.join()Basically, what we are doing is:
1) Telling Pyglet to clear all the OpenGl images on the screen.
2) Start a data-collection thread, which runs in parallel and collects a 1/4 second worth of EEG data.
3) Get the Fourier Transform data + the sums of the values at intervals between 5 and 35 Hertz, calculated in steps. Calculated on the previous batch od EEG data!
4) Create the waterfall image data array, and transfer (blit) the pixels to the framebuffer
5) Use the 'steps' value from (3) to select and blit block images to the framebuffer
6) Create a line graph, and transfer (blit) the pixels to the framebuffer.
7) The image creation and display stuff are done faster than 1/4 of a second, so the 'join()' function makes us wait until the data_thread is finished.
Then Pyglet does its magic and gets everything displayed correctly, and we repeat forever (or the user quits the app).
Waterfall PlotCreation
For the awesome-looking Waterfall plot, we start off by creating a NumPy array:
self.fourier_data = np.zeros((280, 160), dtype=np.int8)The size, (280, 160), will be the final size of the plot in pixels, and the 'dtype' (data type) of the array is an 8-bit integer. Why? Because black and white images can be stored this way, with 255 as white and 0 as black, with the shades of grey in-between.
To update the image, we will:
1) Move all the NumPy pixels up one line, so we have space at the bottom for new data
2) We will call a Fourier transform on the captured EEG data.
3) Add the new data, scaled from 0 to 255 to the near the bottom of the array.
4) Calculate which frequency had the highest amplitude
6) Create two empty (all zeros) row on the bottom of the array, and fill and 4 bright (255's) and the location of the highest amplitude. This will be the marker for the dominant brain wave frequency.
EEG Data PlotImage Creation
Now that you know how the Waterfall plot was created, it's easy to see how we can create a quick plot of the EEG data. We start with an empty NumPy array (all zeros), and column-for-column, we set one pixel to 255 (white) at a vertical height proportional to the EEG data value!
For both the EEG Data Plot and Waterfall Plot, we can use the Pyglet function 'pyglet.image.ImageData' to convert the NumPy arrays to image-data arrays to be blitted to the framebuffer.
And that's it for creating real-time graphics from data using Python and Pyglet! I hope you're inspired to move beyond Matplotlib :)
Controlling Hardware with your MindThe Jetson Nano comes with some fantastic and easy to use hardware interfacing libraries. Our goal is to analyze our brainwaves through the Autoencoder, then use the compressed vector to train a k-mean clustering algorithm, and use the classification results to control physical hardware!
The code can be found in the third and final Jupyter Notebook on Github. For this example, we'll control two LEDs, one green and one red, using values output from a trained k-mean model which is fed Autoencoder data. We can use this as a biofeedback mechanism to learn to control the LED with our minds.
If you need information on how to interface GPIO pins with LED's, I can recommend this page. We aim to have two LEDs set up as described there:
After going through my electronics and not finding the correct jumper wires, I had to join male-to-male and female-to-female wires I had laying around. You should, of course, be using male-to-female wires from your breadboard to your Jetson Nano, and definitely use a red wire for 5 V; black for ground, and light colors for your signal wires! Here's how it looked with my Jetson Nano:
Jetson.GPIO
We can load and test the GPIO library with these python commands:
>>> import Jetson.GPIO as GPIO
>>> print(GPIO.JETSON_INFO)
{'P1_REVISION': 1, 'RAM': '4096M', 'REVISION': 'Unknown', 'TYPE': 'Jetson Nano', 'MANUFACTURER': 'NVIDIA', 'PROCESSOR': 'ARM A57'}As we want to turn our LED's on and off, our control GPIO pins have to be set up as outputs. We will use pins 11 and 12, as they happen to be close to the 5V and ground pins. The command to use these pins as outputs is:
GPIO.setmode(GPIO.BOARD)
GPIO.setup([11, 12], GPIO.OUT)We can test our circuit with:
GPIO.output([11, 12], GPIO.HIGH)which should turn both LEDs on. IF that didn't happen, check your circuit making especially sure that the LEDs are in the correct orientation! Try turning them off again with:
GPIO.output([11, 12], GPIO.LOW)With everything set up, the system should look like this:
Training the Classifier
After we load up the Autoencoder Model, and the various functions we need to read in EEG data and filter it, we can start to classify data to control our LEDs.
We have two cells in the notebook you can use to do this, each looks like this:
state_1 = np.zeros((20, 100))
for i in range(100):
data = process_eeg_data()
state_1[:,i] = data.cpu().numpy()As each capture takes 250 milliseconds, this is 25 seconds of data we can record in a particular brain state. You can try concentrating, reading, listening to music, etc, etc.
Do the same for 'state_2', with a different mindset! Then we can train our k-means algorithm (described above) to learn to differentiate the states! The final code that brings everything together is pretty simple.
while True:
autoencoder_output = process_eeg_data().cpu().numpy()
state = kmeans.predict(autoencoder_output)
if state == 0:
GPIO.output(11, GPIO.HIGH)
GPIO.output(12, GPIO.LOW)
else:
GPIO.output(12, GPIO.HIGH)
GPIO.output(11, GPIO.LOW)And here's how it looks in action:
There you have it! I could reliably control the LEDs alternating between concentrating and chilling out. Of course, you can modify the code to learn to differentiate even more brain simultaneously with k-means, just do more training! Imagine adding some servo motors and having a controllable robot arm, or quadcopter!
SUMMARYYou have learned how to read in and signal process brain-waves. Then how to build and train an autoencoder to compress the EEG data to a latent representation. Then you have used the k-means machine learning algorithm to classify the data to determine brain-state, and used the information to control physical hardware!
And along the way pick up tips on creating GUIs and real-time graphics in Python!
I hope you have enjoyed this tutorial and can see the potential that on-the-edge Artificial Intelligence promises! I look forward to helping out with any questions you might have!











Comments
Please log in or sign up to comment.