My project focuses on predicting the lowest energy levels of molecules using machine learning, combined with 3D molecular visualization through quantum chemistry. I decided to create this project to bridge the gap between quantum mechanics and classical machine learning, making complex molecular analyses more accessible and comprehensible. The project works by first using quantum computing to simulate molecular energy levels and then training a machine learning model on these results to predict energy levels for new molecular configurations. Additionally, it employs 3D visualization tools to display molecular structures, providing an intuitive understanding of molecular geometry and behavior.
Working of my project:
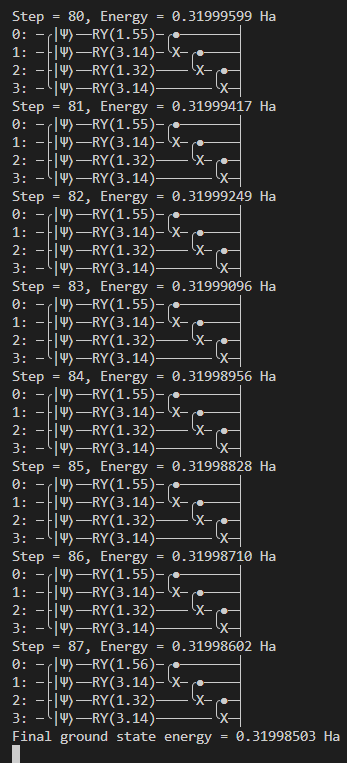
This sample code demonstrates how to use variational quantum eigensolver (VQE) to predict the lowest energy level (ground state energy) of an H2 molecule. The code defines the H2 molecule with a bond length of 0.74 Å, generates its Hamiltonian, and sets up a quantum device to simulate the molecule. The ansatz function builds a variational quantum circuit, and the cost_fn function calculates the energy expectation value of the Hamiltonian using this circuit. The optimization loop iteratively adjusts the circuit parameters to minimize the energy, using a gradient descent optimizer.
To integrate this with a machine learning model, you can extend the dataset by running this quantum simulation for different molecular configurations and bond lengths, collecting the corresponding ground state energies. This dataset can then be used to train a machine learning model to predict ground state energies for new configurations.
For 3D visualization, you can use the coordinates of the molecules and libraries like RDKit or Py3Dmol to visualize the molecular structure. By combining the energy prediction model with the 3D visualization, you create a comprehensive tool that not only predicts molecular properties but also provides an intuitive graphical representation of the molecule.



Comments
Please log in or sign up to comment.