Before the detailed documentation, I have attached the demovideo of my project in below YouTube link, Kindly watch it.Increase in fatality due to Covid-19 in second wave
In second wave, most of the highly populated countries got affected a lot. These are the major reasons for high fallibility rate.
•Doctors/expertise are fully occupied to take care of admitted patients.
•In a highly populated countries, To analyze the data received from IoT medical device need more human power.
•Assessing the home quarantine patients in rural area during pandemic is tedious process due to lack of internet facility.
•Classifying the patients severity takes more time and need more bed occupancy to analyze the patient status.
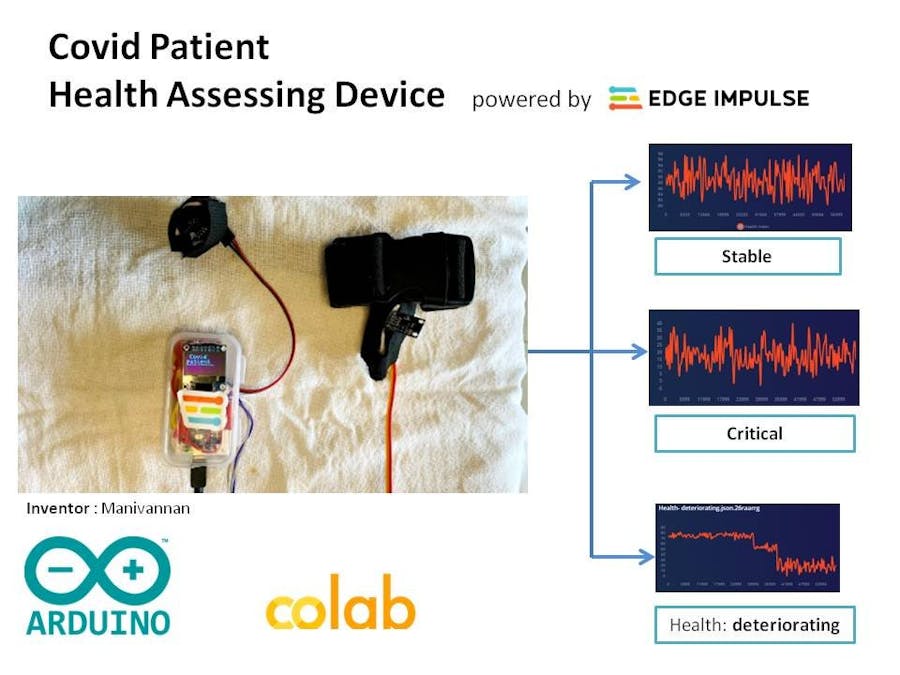
Technology contribution to solve the pandemic problem :•Covid Patient health Assessing device powered by Edge Impulse will analyze the SpO2, heart rate, body temperature and respiratory Rate of a patient.
•The TinyML model is trained by the datasets suggested by medical guidelines.
•Latency is lowest compared to IoT devices
•A 15Kb Rom – health Assessing TinyML model can run on any TinyML supported microcontrollers.
•The device will analyze the health status and classify into Stable, Critical, health : deteriorating
Architecture:The Covid Patent health assessing device involves :
- Health data sets generation using google colab
- Data acquisition in Edge Impulse
- Model training in Edge Impulse
- Real time sensor integration
- model deployment
I have referred the datasets from below research papers and links.
In the below chart, Score 0 Indicates Stable, Score 1 indicates mild, score 2 indicates moderate and Score 3 indicates critical.
The score 0 means stable person with oxygen level >96 where the score 3 indicates oxygen level < 90 and need ICU support.
I have developed a algorithm to calculate the heal index based on SpO2, respiratory rate, heart rate and body temperature.
The SpO2 have higher weightage (40), RR (30), Heart rate (20) and temperature (10).The reason for allocating higher weightage to SpO2 is that the SpO2 health parameter is more crucial in identifying the patient health condition. And also the risk factor is 2 for SpO2.
The health Index will be calculated on below formula:
Health Index = ((40-((100-spo2)*2))+(30-((RR-CAL_RR)*1.5))+(20-(CAL_PULSE-pulse))+(10-((Temperature-CAL_TEMP))));I have created a datasets for SpO2, RR, HeartRate and temperature for below conditions:
- Stable
- Mild
- Moderate
- Critical
kindly use the below GitHub link to create datasets.
https://github.com/Manivannan-maker/CovidPatientHealhAssessingDevice/blob/main/Healthdata.ipynb
Run the code in google colab from your google account login.
The datasets will be saved in Drive folder as.csv format.
Data Acquisition :Single datasets require 3 hours of data for this project. so for 40 datasets, it will take more than 120 hours of effort to make it simple, I have approached alternative method.
The data acquisition works based on below flow:
I have copied the data buffer from google colab into the Arduino and the data selection is based on potentiometer position.
Based on the tuning of potentiometer, the health index algorithm will take input from different datasets ( stable, mild, moderate and critical). I have attached the data acquisition code with goggle colab datasets as array buffer.
Input: 170 Mins health Index Data
Algorithm will convert 170 mins into 59 secs data time frame.
Output : Health index data in 500ms interference delay.
Data forwarding in Edge Impulse :
I have used Data forwarding commands to upload the datasets to the edge impulse. Please refer the below documentation.
https://docs.edgeimpulse.com/docs/cli-data-forwarder
$ edge-impulse-data-forwarderName the label as 'Health Index'.
Datasets: Health- Deteriorating:
Health - stable:
health - critical:
Create Impulse:
NN Classifier:
In NN classifier, Edit in expert mode (keras).
import tensorflow as tf
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, InputLayer, Dropout, Conv1D, Conv2D, Flatten, Reshape, MaxPooling1D, MaxPooling2D, BatchNormalization
from tensorflow.keras.optimizers import Adam
sys.path.append('./resources/libraries')
import ei_tensorflow.training
# model architecture
model = Sequential()
model.add(Flatten())
model.add(Dense(32,
activity_regularizer=tf.keras.regularizers.l1(0.00001)))
model.add(Dense(16,
activity_regularizer=tf.keras.regularizers.l1(0.00001)))
model.add(Dense(8,
activity_regularizer=tf.keras.regularizers.l1(0.00001)))
model.add(Dense(4,
activity_regularizer=tf.keras.regularizers.l1(0.00001)))
model.add(Dense(classes, activation='softmax', name='y_pred'))
# this controls the learning rate
opt = Adam(lr=0.0005, beta_1=0.9, beta_2=0.999)
# this controls the batch size, or you can manipulate the tf.data.Dataset objects yourself
BATCH_SIZE = 32
train_dataset = train_dataset.batch(BATCH_SIZE, drop_remainder=False)
validation_dataset = validation_dataset.batch(BATCH_SIZE, drop_remainder=False)
callbacks.append(BatchLoggerCallback(BATCH_SIZE, train_sample_count))
# train the neural network
model.compile(loss='categorical_crossentropy', optimizer=opt, metrics=['accuracy'])
model.fit(train_dataset, epochs=500, validation_data=validation_dataset, verbose=2, callbacks=callbacks)I have removed 'activation= Relu' in all neural networks since the data is always positive value.
The neural layer is formed as below:
Model Training:
Model Deployment:
Select the Arduino in deployment and built it. the zip file of your project is getting downloaded.
Please make sure to include the library in Arduino.
code integration:After adding the zip file into library, download the below application code from GitHub. The application code will contain SSD1306, Max30102 interface features.
Disclaimer:This project is a proof of concept based on guidelines and research papers. Please do not test directly in patients without Government /medical certifications approval.
I developed this prototype to prepare us against third wave.I believe humankind will overcome this pandemic soon.















_1x_bGT19vVAby.png?auto=compress%2Cformat&w=40&h=40&fit=fillmax&bg=fff&dpr=2)
_3u05Tpwasz.png?auto=compress%2Cformat&w=40&h=40&fit=fillmax&bg=fff&dpr=2)


Comments