The main motive of this project is to create brain machine interface that can be used to address issues faced by people due to limited physical movements of body organs.
My initial idea was to use OpenBCI head mount with 16 electrodes coupled with Ultra96v2, but due to recently imposed import scrutiny I had to opt for a solution that was already available in my country.
So, as an alternative I settled for brainlink lite, which was readily available with local reseller. This is a single electrode device capable of capturing EEG, EMG signals, transmit the same over Bluetooth and it's battery powered!
I tried connecting Brainlink lite to my android phone and use multiple android apps available to infer the data sent from the device. Although connecting to android phone worked without any problem, android apps seemed pretty useless as there was inaccurate outcome of action. Apps consistently failed to determine whether my mind was focused/meditating/distracted.
So I decided, as a PoC, use raw data from Brainlink lite and classify whether a person wearing headband is meditating or not. This will help people like me who cannot meditate for long due to drifts.
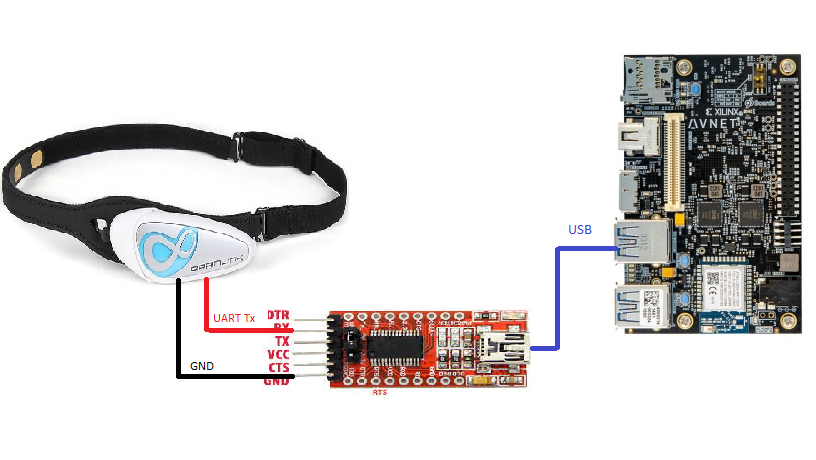
Step 1:First step is to connect Brainlink lite to Ultra96v2. As mentioned earlier, connecting to an Android phone worked seamlessly, but the device failed to connected to Windows and Linux machines. I tried with couple of Windows machines along with Linux running on x86, Ultra96v2 and RPi 3, 4. The issue seem to be with BT connection establishment. So I decided to hack the Brainlink lite!
Hacking begins.....The idea is to connect Brainlink lite over UART to Ultra96v2. First thing I did was open the main module. Below is the image after disassembly. Note two tiny screws in left enclosure.
After some research, it is quite evident that this module used famous Neurosky TGAM chipset which outputs data over UART lines which means Bluetooth module receives data over UART and sends it over radio link. All I need is UART data and GND pin on this module. Below is the close-up shot of BT module and Neurosky chipset.
I started probing BT module's pads for UART Tx using my trustworthy Saleae Logic analyzer. After a few hours, I found the pin on BT module which received UART data from Neurosky chipset. Sampled data would like something like below,
Next step is to solder two wires for UART Tx and GND as done is below image.
Step 2: Brainlink lite output data formatand interfacing with Python
Brainlink lite outputs data in multiple formats. For this project's use case, only raw data will be used. Below is the format in which Brainlink lite outputs raw data,
More on communication protocol: http://developer.neurosky.com/docs/doku.php?id=thinkgear_communications_protocol.Brainlink lite outputs 512 samples per second and the acquisition period in this project will be of 1 second. Below is the Python code excerpt to fetch the EEG data,
sample_count = 0
samples = []
while sample_count < nsamples:
if ReadOneByte(ser) == 0xAA: # Sync 1
if ReadOneByte(ser) == 0xAA: # Sync 2
plen = ReadOneByte(ser) # Packet length
if plen < 169:
if ReadOneByte(ser) == 0x80: # Check for RAW data
if ReadOneByte(ser) == 0x02: # Check the Raw data len
raw = ReadOneByte(ser) * 256 # Read higher 8-bits
raw += ReadOneByte(ser) # Read lower 8-bits
if raw > 32768:
raw -= 65536
samples.append(raw)
sample_count += 1
else:
for i in range(plen): # Other than raw data
ReadOneByte(ser)These samples are then passed onto bandpass filter to allow frequencies between 2Hz and 35Hz after normalizing.
# Compute mean and standard deviation
mean, std = np.mean(samples), np.std(samples)
samples = samples - mean
samples = samples/std
# Filter raw data
filtered_samples = butter_bandpass_filter(samples, lowcut, highcut, fs, order=6)Plotting the filtered and unfiltered raw EEG samples would look like below,
The github repo link provided in code section has a file mindlink.py which does gathering the data and filtering the same. This is used in training and inference stages to fetch data from brainlink lite device.
Below is the code to store training data. For simplicity sake, I have merged both meditating and distracted state data acquisition. Time period is seconds as we are fetching 512 samples and as explained earlier Brainlink lite outputs 512 samples per second. First we meditate and collect data and then collect data while in distracted/non-meditating state.
import numpy as np
import sys
from serial import Serial
from mindlink import read_raw_eeg
from random import shuffle
def main():
eeg_data = []
com_port = '/dev/ttyUSB0'
x_filename = 'x.npy'
y_filename = 'y.npy'
period_in_seconds = 500
ser = Serial(com_port, 57600, timeout=None)
training_data = []
# First collect meditation data
i = 0
label = 1 # For meditation
while i < period_in_seconds:
i += 1
sample, _ = read_raw_eeg(ser, 512)
training_data.append([label, sample])
# Next collect distraction data
i = 0
label = 1 # For distraction
while i < period_in_seconds:
i += 1
sample, _ = read_raw_eeg(ser, 512)
training_data.append([label, sample])
# Shuffle the data
shuffle(training_data)
x_data = []
y_data = []
for label, features in training_data:
x_data.append(features)
y_data.append(label)
# save as numpy array
eeg_data = np.array(x_data)
np.save(x_filename, x_data)
eeg_data = np.array(y_data)
np.save(y_filename, y_data)
if __name__ == "__main__":
main()Time period can be reduced as per one's ability to meditate for longer period of time. After acquisition, this acquired data is shuffled along with labels for randomization during training and stored as numpy array for data
Step 4: Preping the hardwareAs I would be needing a bandpass filter, my obvious choice was to use PYNQ image as it provides support for DPU along hardware accelerated digital filters using python packages.
Download PYNQ for Ultra96v2 from here. Although v2.6 is available currently, v2.5 is used for this project assuming this release would have matured in terms of workaround/solution for known/unknown issues :).
Flash the downloaded image on to 16GB micro SD card using any imager. I used win32diskimager. Next, boot the device with micro SD card inserted. Login credentials is xilinx:xilinx. Python packages pyserial, scipy would be needed for this project, so let's go ahead and install those using below command. These packages should be installed as superuser. Pass for superuser is 'xilinx'.
su
pip3 install pyserial scipyNext, in the requirement list in support for DPU. Clone Xilinx's DPU PYNQ repo as superuser,
git clone --recursive --shallow-submodules https://github.com/Xilinx/DPU-PYNQ.gitIssue make command to initiate the build,
cd DPU-PYNQ/upgrade
makeThis is the process to be followed to upgrade PYNQ 2.5 to be DPU ready.
Next, install 'pynq-dpu' python package using below command,
pip3 install pynq-dpuAfter successful installation, time to test DPU by loading bit file.
root@pynq:/home/xilinx# python3
Python 3.6.5 (default, Apr 1 2018, 05:46:30)
[GCC 7.3.0] on linux
Type "help", "copyright", "credits" or "license" for more information.
>>> from pynq_dpu import DpuOverlay
>>> overlay = DpuOverlay("dpu.bit")
>>>
root@pynq:/home/xilinx# dexplorer -w
[DPU IP Spec]
IP Timestamp : 2020-03-26 13:30:00
DPU Core Count : 1
[DPU Core Configuration List]
DPU Core : #0
DPU Enabled : Yes
DPU Arch : B1600
DPU Target Version : v1.4.1
DPU Freqency : 300 MHz
Ram Usage : Low
DepthwiseConv : Enabled
DepthwiseConv+Relu6 : Enabled
Conv+Leakyrelu : Enabled
Conv+Relu6 : Enabled
Channel Augmentation : Disabled
Average Pool : EnabledOptionally, PYNQ-DPU jupyter notebooks can be downloaded using below command,
cd $PYNQ_JUPYTER_NOTEBOOKS
pynq get-notebooks pynq-dpu -p .These jupyter notebooks can be run on web-browser by accessing following URLs in address bar,
- over SSH: <device_ip>:9090/notebooks/pynq-dpu
- over USB: 192.168.3.1:9090/notebooks/pynq-dpu
- Native: localhost:9090/notebooks/pynq-dpu
Passphrase at login screen is 'xilinx'. Click on suitable example and 'run' cells to get the expected output.
Step 5: Training and graph conversionI'm using Ubuntu 18.04 running on Intel Core-I7 8086K.
First install docker needed for infrastructure around Vitis AI using command,
sudo apt install docker.ioEnsure linux user is in docker group by following instructions provided here.
Pull the latest Vitis repo from github using following command,
git clone --recurse-submodules https://github.com/Xilinx/Vitis-AIOnce checkout is complete, run the docker command to fetch latest docker image,
cd Vitis-AI
./docker_run.sh xilinx/vitis-ai:latestAfter successful pull, time to build docker from recipe. Make sure to exit earlier docker workspace by typing "exit".
cd ./docker
./docker_build_cpu.shOnce the build is complete we can use it according our needs.
For graph conversion we would need hardware handoff for the DPU. This can be obtained from here.
Note: I got the information of which hwh file to use from the PYNQ-DPU repo. A file by name dpu.hwh.link contains hyperlink of hwh file for Ultra96, ZCU104, ZCU111. Below is the content of dpu.hwh.link,
root@pynq:~/proj# cat DPU-PYNQ/pynq_dpu/overlays/dpu.hwh.link
{
"Ultra96": {
"url": "https://www.xilinx.com/bin/public/openDownload?filename=pynqdpu.dpu.ultra96.hwh",
"md5sum": "8e1f936b1e070c6b02c582109d720386"
},
"ZCU104": {
"url": "https://www.xilinx.com/bin/public/openDownload?filename=pynqdpu.dpu.zcu104.hwh",
"md5sum": "d7edbaa5e2a65234a89eb8e50e35f6fc"
},
"ZCU111": {
"url": "https://www.xilinx.com/bin/public/openDownload?filename=pynqdpu.dpu.zcu111.hwh",
"md5sum": "acefbbcb547ef9c84dcd992e629ee87c"
}
}root@pynq:~/proj#The github repo for this project is available here. The repo consists of the following files,
- docker_run.sh: Top level script to start docker session.
- image_input_fn.py: Python file to create a list of training data needed for quantization.
- train_keras.py: Python file used to create a neural network graph and perform training.
- keras_2_tf.py: File from Xilinx to convert keras out h5 file to tensorflow checkpoint files.
- x.npy: Training data in numpy format containing raw data from Brainlink lite.
- y.npy: Label array corresponding to data present in x.npy.
- run.sh: Toplevel script to perform training and graph conversion.
- pynqdpu.dpu.ultra96.hwh: Hardware handover file explained earlier.
- dpu-03-26-2020-13-30.dcf: File generated using command "dlet -f <hwh_file>.
- ULTRA96V2.json: json architecture file used for model compilation.
- target: This is the folder consisting of file that can be run on Ultra96v2.
- target/dpu_bam.elf: Converted model obtained after successfully running "run.sh".
- target/app_perf.py: Python3 script to measure performance on Ultra96v2 using generated model.
- app.py: Actual Python3 script to perform inference on real-time input and measure meditation level.
- mindlink.py: Python3 file to fetch the data from Brainlink lite using serial port and pass it through bandpass filter.
In order to simply deployment, a top-level script is used to define parameters required, perform training, freeze the graph, quantize the graph and finally convert the graph.
How to train and generate model?Clone the git repo using command,
git clone https://github.com/NitinBhaskar/BAM.gitNext, 'cd' to the checked-out repo and start the docker,
cd BAM
sudo ./docker_run.sh xilinx/vitis-ai:latestAccept the 'terms' of usage and issue following command,
source run.shThis top-level script activates Tensorflow workflow, initializes the parameters, creates and trains the graph, converts h5 to tensorflow checkpoint files, freezes the checkpoint files, quantizes the frozen model, compiles the model and finally copies required files to 'target' folder.
Let's go through this top-level script section by section.
First, activate tensorflow workflow,
conda activate vitis-ai-tensorflowVarious parameters are initialized next,
export OUT=./out
export CHKPT_FILENAME='chkpt.ckpt'
export GRAPH_NAME='model.pb'
export FROZEN_GRAPH=frozen_model.pb
export TARGET_DIR=./target
export ARCH=/workspace/ULTRA96V2.json
export INPUT_HEIGHT=32
export INPUT_WIDTH=16
export INPUT_CHAN=1
export INPUT_SHAPE=?,${INPUT_HEIGHT},${INPUT_WIDTH},${INPUT_CHAN}
export INPUT_NODE=conv2d_input
export OUTPUT_NODE=dense/BiasAdd
export NET_NAME=bamAlthough, we get single dimensional output from Brainlink lite, we reshape the same to INPUT_SHAPE=(?, 32, 16, 1) due to hard requirement of graph conversion infrastructure which does not support single dimensional flat input. Various other parameters such as output directory, checkpoint and graph filenames along with architecture json file name with input and output node names are defined too.
Next, create output directory and start the training,
mkdir -p ${OUT}
echo "****************************** training *******************************"
# Start the training
python train_keras.pytrain_keras.py contains neural network model that is defined as below,
model = tf.keras.Sequential([
tf.keras.layers.Input(train_x.shape[1:]),
tf.keras.layers.Conv2D(32, kernel_size=(2, 2), activation="relu"),
tf.keras.layers.MaxPooling2D(pool_size=(2, 2)),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(num_classes)
])model.summary() yields below output,
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
conv2d (Conv2D) (None, 31, 15, 32) 160
_________________________________________________________________
max_pooling2d (MaxPooling2D) (None, 15, 7, 32) 0
_________________________________________________________________
flatten (Flatten) (None, 3360) 0
_________________________________________________________________
dense (Dense) (None, 2) 6722
=================================================================
Total params: 6,882
Trainable params: 6,882
Non-trainable params: 0Training and test data are obtained from x.npy and y.npy files. After reading data and label files, these list are split to 80% training data and 20% test data as below and reshape to INPUT_SHAPE as explained earlier,
x_data = np.load('x.npy')
y_data = np.load('y.npy')
l = int(len(x_data) * 0.8)
train_x = np.asarray(x_data[:l])
train_y = np.asarray(y_data[:l])
test_x = np.asarray(x_data[l:])
test_y = np.asarray(y_data[l:])
train_x = train_x.reshape(-1, INPUT_HEIGHT, INPUT_WIDTH, INPUT_CHAN)
test_x = test_x.reshape(-1, INPUT_HEIGHT, INPUT_WIDTH, INPUT_CHAN)Next is to compile, fit, evaluate and save the model,
model.compile(loss='categorical_crossentropy',
optimizer=tf.keras.optimizers.Adam(lr=LEARN_RATE, decay=DECAY_RATE),
metrics=['accuracy']
)
model.fit(train_x, train_y, epochs=10)
test_loss, test_acc = model.evaluate(test_x, test_y, verbose=2)
print('\nTest accuracy:', test_acc)
model.save(os.path.join(OUT,'keras_out.h5'))After completing 10 Epochs, we can see below result,
Epoch 10/10
2000/2000 [==============================] - 0s 156us/sample - loss: 0.6931 - acc: 0.7520
WARNING:tensorflow:From train_keras.py:44: The name tf.keras.backend.get_session is deprecated. Please use tf.compat.v1.keras.backend.get_session instead.
500/500 - 0s - loss: 0.6931 - acc: 0.7280
Test accuracy: 0.728Training accuracy of 75.20% and test accuracy of 72.8% is obtained with the data and model defined.
Next, keras_2_tf.py converts generated h5 to tensorflow checkpoint files,
python keras_2_tf.py --keras_hdf5 ${OUT}/keras_out.h5 \
--tf_ckpt=${OUT}/tf_chkpt.ckptThis python script print the input and output nodes that is required during quantization and model compilation.
Keras model information:
Input names : [<tf.Tensor 'conv2d_input:0' shape=(?, 32, 16, 1) dtype=float32>]
Output names: [<tf.Tensor 'dense/BiasAdd:0' shape=(?, 2) dtype=float32>]Next stage is quantization, which takes '100' calibration iterations mentioned using parameter '--calib_iter 100' in run.sh. Output of quantization phase is as below,
INFO: Calibration Done.
INFO: Generating Deploy Model...
INFO: Deploy Model Generated.
********************* Quantization Summary *********************
INFO: Output:
quantize_eval_model: ./out/quantize_eval_model.pb
deploy_model: ./out/deploy_model.pbNext comes model compilation which takes quantized graph and architecture information as input and generates an elf file that can be used for inference on Ultra96v2.
The architecture file ULTRA96V2.json is as below,
{
"target" : "DPUCZDX8G",
"dcf" : "/workspace/dpu-03-26-2020-13-30.dcf",
"cpu_arch" : "arm64"
}As explained earlier dpu-03-26-2020-13-30.dcf is obtained by running dlet on pynqdpu.dpu.ultra96.hwh. After compilation is successful, kernel list for neural network is printed as shown below.
****************************** compiling *********************************
**************************************************
* VITIS_AI Compilation - Xilinx Inc.
**************************************************
Kernel topology "bam_kernel_graph.jpg" for network "bam"
kernel list info for network "bam"
Kernel ID : Name
0 : bam
Kernel Name : bam
--------------------------------------------------------------------------------
Kernel Type : DPUKernel
Code Size : 2.01KB
Param Size : 6.72KB
Workload MACs : 0.13MOPS
IO Memory Space : 3.78KB
Mean Value : 0, 0, 0,
Total Tensor Count : 3
Boundary Input Tensor(s) (H*W*C)
conv2d_input:0(0) : 32*16*1
Boundary Output Tensor(s) (H*W*C)
dense_MatMul:0(0) : 1*1*2
Total Node Count : 2
Input Node(s) (H*W*C)
conv2d_Conv2D(0) : 32*16*1
Output Node(s) (H*W*C)
dense_MatMul(0) : 1*1*2Finally, generated elf file is copied into 'target' folder.
Step 6: DeployingOnce the graph is ready for deployment after completing above described process, SCP target folder to Ultra96v2 using following command,
scp -r target xilinx@<ultra96v2_IP>://home/xilinx/Change the privileges to superuser by issuing 'su' command. There are two 'app's which are python scripts that can be used to perform inference using the graph.
First is app_perf.py which can be used to measure the performance of the generated and quantized graph. This script takes entire training and test data that were used earlier and runs inference on each one of them and then computes the accuracy by matching actual label with predicted label.
Second is app.py which take real-time data from brainlink lite and perform inference and measure meditation level and displays the same after time_period amount of time.
Both the examples have few things in common such as loading DPU bit file and converted graph,
from pynq_dpu import DpuOverlay
overlay = DpuOverlay("dpu.bit")
overlay.set_runtime("vart")
overlay.load_model("dpu_bam.elf")Next is initializing input/output parameters based on graph,
dpu = overlay.runner
inputTensors = dpu.get_input_tensors()
outputTensors = dpu.get_output_tensors()
tensorformat = dpu.get_tensor_format()
if tensorformat == dpu.TensorFormat.NCHW:
outputHeight = outputTensors[0].dims[2]
outputWidth = outputTensors[0].dims[3]
outputChannel = outputTensors[0].dims[1]
elif tensorformat == dpu.TensorFormat.NHWC:
outputHeight = outputTensors[0].dims[1]
outputWidth = outputTensors[0].dims[2]
outputChannel = outputTensors[0].dims[3]
else:
raise ValueError("Image format error.")
outputSize = outputHeight*outputWidth*outputChannel
shape_in = (1,) + tuple(
[inputTensors[0].dims[i] for i in range(inputTensors[0].ndims)][1:])
shape_out=(1,outputHeight,outputWidth,outputChannel)Preparing input data and output placeholders,
input_data = []
output_data = []
input_data.append(np.empty((shape_in),
dtype = np.float32, order = 'C'))
output_data.append(np.empty((shape_out),
dtype = np.float32, order = 'C'))
inData = input_data[0]
inData[0,...] = in_q.reshape(
inputTensors[0].dims[1],
inputTensors[0].dims[2],
inputTensors[0].dims[3])Invoking DPU to perform inference,
job_id = dpu.execute_async(input_data, output_data)
dpu.wait(job_id)Below is the output seen for app_perf.py.
root@pynq:/home/xilinx/target# python3 app_perf.py
Correct: 1850 Wrong: 650 Accuracy: 0.74As it can be see the accuracy is 74% which is between test accuracy(72.8%) and training accuracy(75.2%) as mentioned above which is very good considering quantization did not result in significant loss in accuracy.
The main python app to obtain meditation level is 'app.py' which can be invoked using below command,
python3 app.py -m dpu_bam.elf -s <com_port> -t <time_period>app.py does things bit differently than app_perf.py.
Two threads are created 1. 'Producer thread' - To read out the raw data from Brainlink lite via serial port, 2. 'Consumer' thread - To consume read out data and pass it to DPU to predict whether the person is currently in meditation state. This way both threads can run concurrently without waiting on other thread to finish.
This app first initializes Queues and serial port. Queue 'q' is used to synchronize between 'producer' and 'consumer' threads. 'common_q' is used to post message from main thread to 'producer' thread and from 'consumer' thread to main thread. This way exclusivity is maintained among 3 threads.
q = Queue() # Queue to share EEF raw data
common_q = Queue() # Queue to sync between main, producer and consumer threads
time_period = int(t_in_sec) # Time to run in seconds
ser=Serial(com_port,57600,timeout=None) # Serial port initInitialize and start two threads, but first initialize and start 'producer' thread for it to be ready with data ASAP.
t1 = threading.Thread(target = producer, args =(q, ser, common_q)) # Init thread1
t1.start() # Start the thread
t2 = threading.Thread(target = consumer, args =(q, common_q)) # Init thread2
t2.start() # Start the threadPut time period in common_q which will be consumed by producer thread and forwarded to consumer thread. After posting the time period main thread sleeps for time period amount time and let 'producer' and 'consumer' finish their task and get back with the result.
# Start producer and consumer for t_in_sec seconds
common_q.put(int(t_in_sec))
# Wait till time_period
time.sleep(int(t_in_sec))Next main thread waits till 'consumer' posts meditation level to common_q and then prints out the meditation level percentage.
out = common_q.get() # Read meditation level value
print('Meditation level was %.2f%%'%(out/int(t_in_sec)*100))Finally wait till all the threads exit gracefully,
t1.join()
t2.join()Below is the demo where I am trying to meditate and every second state is printed whether I am meditating or distracted. After the completing the time period the result is printed as 80% which is in terms of percentage of time I was meditating.
Conclusion:This is a PoC that show what is possible with EEG raw data and good edge device capable of performing inference with good accuracy. This can be extended/enhanced to have multiple electrodes connected as in OpenBCI and with combination of input from multiple electrodes more can be achieved for broader real world use cases.











_3u05Tpwasz.png?auto=compress%2Cformat&w=40&h=40&fit=fillmax&bg=fff&dpr=2)
Comments