AI is becoming an essential part of modern healthcare. The use of ambient AI solutions to reduce clinician burnout from the burdens of documentation is a priority now. AI can guide providers during their patient interactions to prevent errors of omission or underuse, and prevent over-treatment. This increases the probability that providers will succeed in their transition to value-based reimbursement. With the growing applications in the field AI innovations and algorithms associated with the medical image process are evolving rapidly. Accessing the power of Xilinx platforms to solve challenging problems in their domain, accelerate time to insight, and innovate ahead of the curve is the ex-facto to better efficiency and lower latency. The reconfigurability of Xilinx platforms provides longer life utility of their products as well. Xilinx’s full-stack deep learning SDK, Vitis AI, along with highly adaptive Xilinx’s AI platforms, enable medical equipment manufacturers and developers with rapid prototyping of these highly evolving algorithms, minimizing time-to-market and cost.
Medical images having extensive properties (higher resolution, largely electronic and photonic 3D format, etc.), these images require significant preprocessing before feeding them into an AI inference engine. These preprocessing algorithms require that several complex operations be performed on each pixel in each image, resulting in a large number of operations per second. Implementing these preprocessing algorithms in Xilinx platforms along with the Xilinx’s AI inference engine (DPUs) provides near real-time detection, which can be essential in medical care. Xilinx’s Vitis AI provides an end-to-end development platform for these whole application acceleration solutions.
Acute otitis media (AOM) is a painful type of ear infection. It occurs when the area behind the eardrum called the middle ear becomes inflamed and infected. Otitis Media is generally caused by viruses or bacteria, often as a complication of the common cold or allergies. Although acute otitis media can occur at any age, it is most common between the ages of 3 months and 3 years. Despite advances in the development of pneumococcal conjugate vaccines, acute otitis media (AOM) is a common childhood infection, caused mainly by Streptococcus pneumoniae. It has been suggested that the persistence of pneumococcal nasopharyngeal carriage is a risk factor for subsequent recurrent infections. With an increase in respiratory tract infections during Covid-19, complications of Otitis media is surging. Otitis media not only causes severe pain but may result in serious complications if it is not treated. An untreated infection can travel from the middle ear to the nearby parts of the head, including the brain and lead to infective meningitis. Signs and symptoms differ with age groups and generally grouped under the following:
- Neonates: Irritability or feeding difficulties may be the only indication of a septic focus.
- Older children: This age group begins to demonstrate a consistent presence of fever and otalgia, or ear tugging
- Older children and adults: Hearing loss becomes a constant feature of AOM and otitis media with effusion (OME)
The financial burden of eardrum diseases to society is enormous; for example, more than $5 billion per year is spent on acute otitis media (OM) alone** because of unnecessary antibiotics and the over treatment of it. This contributes to antibiotic resistance as well. The incidence rates of acute OM and chronic OM are 10.9% and 4.8%, respectively, with 51.0% and 22.6% of these occurring in children under the age of five years**. Although, diagnosis for acute otitis media is not difficult and the disease can be easily cured, but because of the complications affecting children’s language learning and cognitive processes, it is essential to diagnose these diseases in a timely and accurate manner.
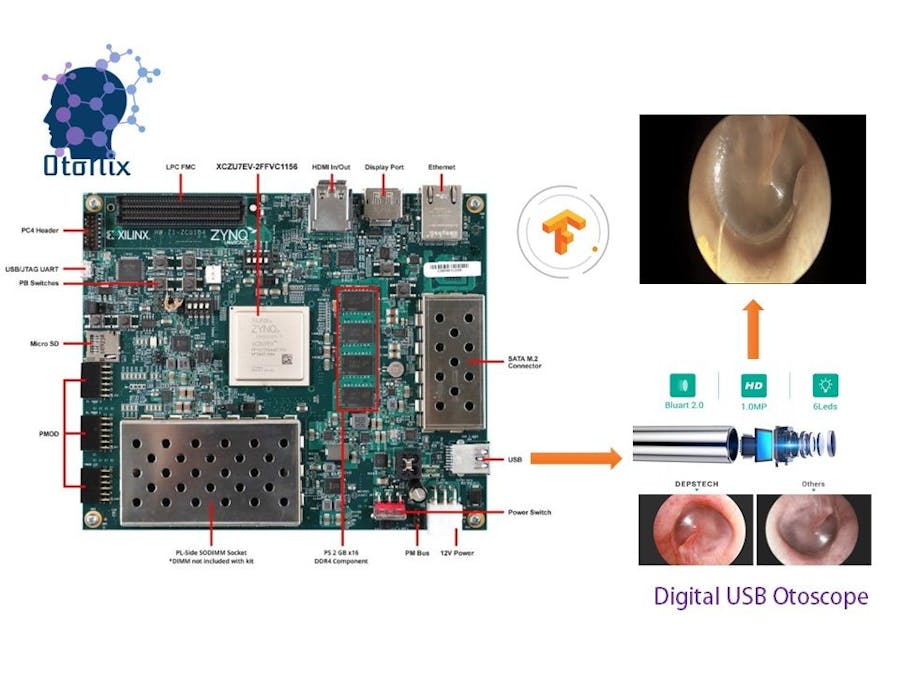
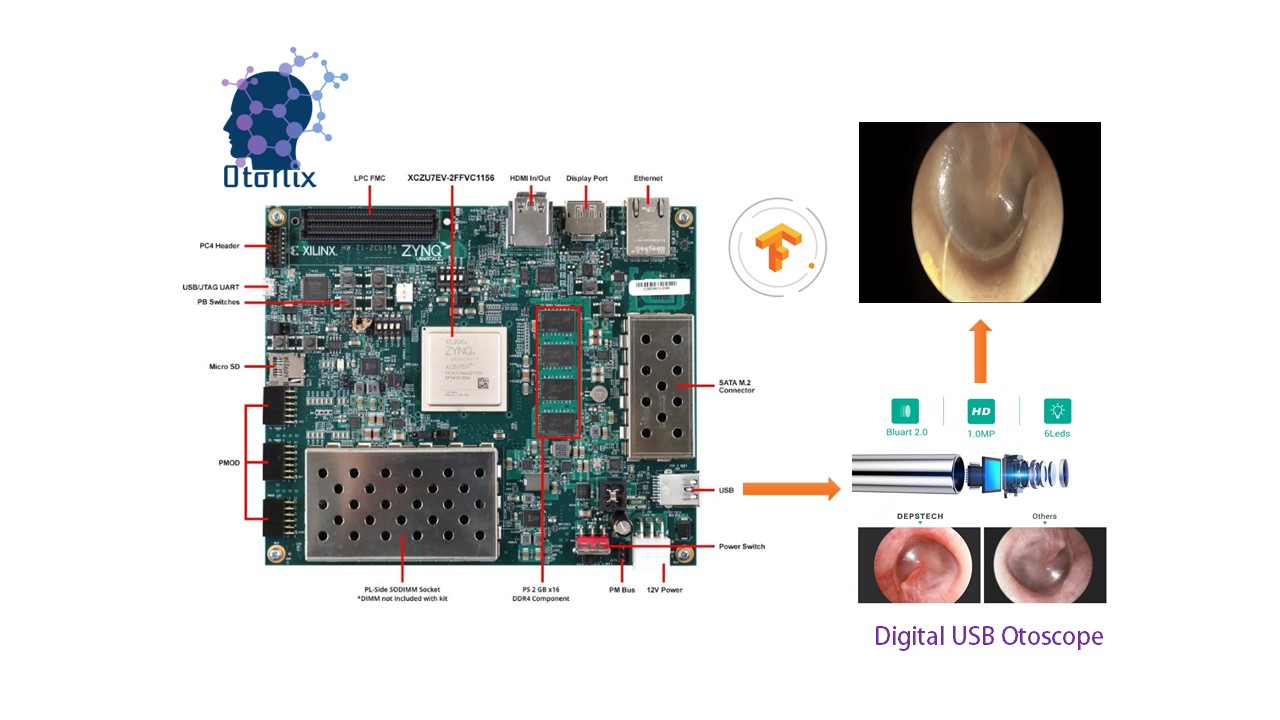
Xilinx OtoNix or Xilinx based Otoscopy Using Neural Imaging is our solution to diagnose AOM and other ear tracts infections. OtoNix is one of its kind to diagnose for tympanic membrane based on image classification techniques.
Our device OtoNix is going to demonstrate how a medical application developer can easily take a medical image dataset and develop and evaluate an end-to-end AI accelerated application using Vitis AI, without writing any lower level RTL code. I took the tympanic membrane otoscopy image dataset with proper labels from here http://www.ctganalysis.com/Content/tympanic-membrane-data-set, then preprocess the images, create a machine learning network and train it on Google Colab platform. Then we use the Xilinx’s Vitis AI toolset to quantize, compile and evaluate the model in one of the Xilinx’s AI inference platforms, the ZCU104 board.
Model Creation and Training:Design Considerations:1. Transfer Learning consideration: Training a deep neural network requires a large dataset. Collecting a large number of medical images is always a challenge. Our tympanic membrane dataset only has 968 images, which is insufficient for training a deep network from scratch. To overcome this challenge, we use a Transfer Learning technique. Transfer Learning can be achieved in two ways, finetuning a network pre-trained on general images or fine-tuning a network pre-trained on medical images for a different target organ or task. For ease of availability, we chose to use an Inception v3 network pre-trained with ImageNet, a dataset of over 14 million general images.
2. Dataset size consideration: The size of the dataset is a key parameter in deciding on the level of Transfer Learning. Even with some augmentation, our tympanic membrane dataset is small. The number of parameters in the Inception v3 network is very large, roughly 22 million, so full adaption may result in overfitting. Thus, we chose to proceed with partial adaption.
3. Vitis AI considerations: When creating a deep neural network or choosing one for Transfer Learning targeting Xilinx’s AI platforms for deployment, we need to make sure all layers, the size of the kernels and the activation functions in the network is supported by the Xilinx Vitis AI stack. Our selected Inception v3 for Transfer Learning is fully supported by the Vitis AI stack. Also, in our network modifications both Dense and Softmax layers are supported (more on this coming later). The kernel size and the input/output dimensions of these newly added layers are within the supported range as well. For more details on the supporting layer, kernel size etc. please refer to Xilinx’s DPU IP Product Guide PG338.
Supervised training for our automated otitis media image classification requires homogenous, clean and labelled datasets of images. The dataset we are using is available at http://www.ctganalysis.com/Content/tympanic-membrane-data-set,[The above database is free to use for non-commercial purposes] this database consists of totally 956 otoscope images collected from the eligible patients who were examined at Özel Van Akdamar Hospital in Turkey between 10/2018 and 6/2019:
- The
resolutionof the images in the database is500-by-500 pixels. - The number of
normalsamples is535.
The number of abnormal samples is 421.
- The number of
AOMsamples is119. - The number of
CSOMsamples is63. - The number of
Earwaxsamples is140.
The number of other samples is 99.
- The number of
otitis externais41. - The number of
ear ventilation tubeis16. - The number of
foreign bodiesin the ear is3. - The number of
pseudo-membranesis11. - The number of
tympanosklerossamples is28.
Step 1:Creating metadata of images along with labels
We need to create a.csv file containing all image_id and their associated labels[ this is a good practice because it would be helpful in creating directories for training, validation and test images using python script]. First of all, we only need seven classes or image sets, viz. aom, csom, earwax, ear ventilation tube,otitis externa, tympanoskleros and normal( we are not going to train forthe rest of the classes in the dataset as they have very few images). Follow the steps below to make a.csv file for your own image dataset:
Now let's list all file in our dataset folder directory using cmd on windows:
REM list all filenames in mounted directory
--dir /b
REM list all filenames as well as subfiles with full path names
--dir /b/s
REM command to get all file names and save to .txt file
--dir /b/s/w *.png > "metadata.txt"Using some basic notepad and Excel tricks you can easily store image_id along with labels in a.csv file format.
Step 2: Exporting images from the dataset folder
We have to make separate directories for training, testing and validation, train_test_split( ) function helps us to divide our whole otoscopy dataset metadata proportionately. Also, we have used pandas profiling and sweetviz to explore the dataset.
# Read the metadata
pdf = pd.read_csv('/content/drive/MyDrive/tympanic_membrane_dataset/metadata.csv')
# Explore the dataset using pandas profiling
pp.ProfileReport(pdf)
# Explore the dataset using sweetviz
sweet_report = sv.analyze(pdf)
sweet_report.show_html()Splitting the metadata for training, testing and validation
# Set y as the labels
y = pdf['dx']
# Split the metadata into training and validation
df_train, df_val = train_test_split(pdf, test_size=0.33, random_state=1, stratify=y)
# Print the shape of the training and validation split
print(df_train.shape)
print(df_val.shape)
# Find the number of values in the training and validation set
df_train['dx'].value_counts()
df_val['dx'].value_counts()We have chosen 33% as our test size, and stratify as true to keep the metadata splitting proportionate.
After creating directory let's, transfer all images into them for the final training process. Below is the code for transferring all the images from dataset folder to newly created test, validation and train directories:
# Transfer the training images
for image in train_list:
fname = image + '.png'
label = pdf.loc[image, 'dx']
if fname in folder:
# source path to image
src = os.path.join('/content/drive/MyDrive/tympanic_membrane_dataset/images', fname)
# destination path to image
dst = os.path.join(train_dir, label, fname)
# copy the image from the source to the destination
shutil.copyfile(src, dst)
# Transfer the validation and test/quatizer images
testimgcount=0
#open the testlabelfile
f = open(os.path.join(base_dir, 'testlabelfile.txt'), 'a+')
fcal = open(os.path.join(base_dir, 'calibration.txt'), 'a+')
for image in val_list:
fname = image + '.png'
label = pdf.loc[image, 'dx']
if fname in folder:
# source path to image
src = os.path.join('/content/drive/MyDrive/tympanic_membrane_dataset/images', fname)
# destination path to image
dst = os.path.join(val_dir, label, fname)
# copy the image from the source to the destination
shutil.copyfile(src, dst)
#copy for testing and dnndk quantization
if testimgcount<50:
dst = os.path.join(test_dir,label, fname)
shutil.copyfile(src, dst)
#append the testlabel file
f.write(label+'\n')
#append in the calibration.txt file
imgloc = os.path.join(label,fname)
fcal.write('{0}\n'.format(imgloc))
# increment the file#
testimgcount+=1
f.close()
fcal.close()After transferring the images, we can clearly see the difference in image quantities per class. However, such a large difference in image quantities per class is not good for achieving maximum accuracy, so we are going to augment the images to increase our training image dataset and also to keep images in proportionate quantities. [Image augmentation is a super effective concept when we don’t have enough data with us.]
Step 3: Augmenting Images using KerasImageDataGenerator()
Image augmentation artificially creates training images through different ways of processing or combination of multiple processing, such as random rotation, shifts, shear and flips, etc. Training the network with this dataset as-is would create a model which is heavily skewed by a single majority class, normal. To prevent this, we use an offline data augmentation technique to reduce class imbalance. To create augmented images we use the Keras ImageDataGenerator class as follows,
# Create a data generator to augment the images in real time
datagen = ImageDataGenerator(
rotation_range=180,
width_shift_range=0.1,
height_shift_range=0.1,
zoom_range=0.1,
horizontal_flip=True,
vertical_flip=True,
# brightness_range=(0.9,1.1),
fill_mode='nearest')You can clearly see that after augmenting the images we have obtained slightly better dataset which can now be used for ML training. Also, we need to resize all images to 224 x 224 px to be fed into InceptionV3 network(Many images resize tools are already available for this)
How augmented images look like?
Our objective is to classify ear otoscopic images to seven classes, that are: aom(acute otitis media), csom(chronic suppurative otitis media), earwax, ear ventilation tube, normal, otitis externa, tympanoskleros. We are using Inception v3 network for transfer learning, Inception v3 is a widely-used image recognition model that has been shown to attain greater than 78.1% accuracy on the ImageNet dataset. The model is the culmination of many ideas developed by multiple researchers over the years. It is based on the original paper: "Rethinking the Inception Architecture for Computer Vision" by Szegedy, et. al. Inception V3 network is trained on more than a million images of Imagenet dataset, this pre-trained network can predict up to almost 1000 classes. To meet our application requirement, we augment the network to support classification between 7 output classes. We add a prediction or Softmax layer with seven output classes.
# Create a inception_v3 model alonge with weights
iv3_model = keras.applications.inception_v3.InceptionV3(include_top=False, weights='imagenet', input_shape=(500, 500, 3))
# See a summary of the layers in the model
iv3_model.summary()
# Taking the output of the inception_v3 just before last layer
x = iv3_model.output
# flattening the outputs of the last conv layer
flatten = Flatten()(x)
# adding two fully connected layers. Meeting DPU requirement, keeping output/input ratio at @ 1/6
dense1 = Dense(2048, activation= 'relu')(flatten)
dense3= Dense(128, activation= 'relu')(dense1)
# adding the prediction layer with 'softmax'
predictions = Dense(7, activation='softmax')(dense3)
# Create a new model with the new outputs
model = Model(inputs=iv3_model.input, outputs=predictions)
# See a summary of the new layers in the model
model.summary()The optimum accuracy I got was 87% after I tried several times on Kaggle as well as Google Colab, but after certain epoch, I got Resource Exhausted Error as shown below(my RAM, as well as GPU, was fully used always). But at last, I got success when I made a lesser batch, epoch and learning rate, so I did not get much accuracy. Tweaking the batch size and epoch I was able to convert my model to.pb file via floydhub (floyedhub gives 61GB memory and 200GB SSD free for 2 hours on signup, so memory buffer-overflow was never an issue like I faced on Kaggle. The training file is attached in the code section. However, for demonstration purpose, this step is enough.
Vitis AI quantizer tool accepts.pb format file, also known as frozen graph format. Xilinx provided it's own AI performance and evaluation tool, Vitis AI, the main advantage of using ZCU104 and Vitis AI is a better inference latency, smaller power and memory footprint. To port, the model for Xilinx AI we need to convert or better to say quantize the 32-bit floating-point weights of the trained model into lower precision, as Xilinx DPU(Deep Learning Processing Unit) is a programmable engine which executes highly optimized instruction set. So we need to compile or Inception V3 network into a set of instructions that can be executed by DPU but hang on we don't need so much programming skills here as Xilinx Vitis already has made our work easier by necessary toolchain to quantize and compile via terminal scripts. First, let us load Vitis AI Software via Docker hub, we clone the Vitis AI 1.0 GitHub repository and pull Xilinx/vitis-aitools-1.0.0-CPU Docker image from Docker hub. Step by step instructions is available in the Vitis AI guide UG1414. Here are the command snippets:
sudo systemctl enable docker
# pull the vitis-ai tools image from Docker hub
$ Docker pull xilinx/vitis-ai:tools-1.0.0-cpu
#Clone the Vitis AI repository
$ git clone https://github.com/xilinx/vitis-ai
# Launch the Docker container
$ cd Vitis-AI
$ ./docker_run.sh xilinx/vitis-ai:tools-1.0.0-cpu
# Once inside the container at /workspace, activate the vitis-ai-tensorflow conda
environment
$ conda activate vitis-ai-tensorflowQuantizing the wights, we call vai_q_tensorflow_quantize, for our convenience we have written a script for this task
$ vai_q_tensorflow quantize \
--input_frozen_graph ${TF_NETWORK_PATH}/${FROZEN_MODEL} \
--input_fn ${INPUT_FN} \
--input_nodes ${INPUT_NODES} \
--output_nodes ${OUTPUT_NODES}\
--input_shapes ?,224,224,3 \
--calib_iter 10 \
--method 1 \
--gpu 0 \
--output_dir ${TF_NETWORK_PATH}/qoutputCompilation of the network graph:
The Vitis AI tools Docker comes with Vitis AI VAI_C, a domain-specific compiler. It efficiently maps the network model into a highly optimized instruction sequence for the Xilinx’s Deep learning Processor Unit (DPU). We call the via_c_tensorflow:
# deploy_model.pb is the fiel we obtained in qunatization step
$ vai_c_tensorflow --frozen_pb qoutput/deploy_model.pb \
--arch {ARCH} \
--output_dir coutput/output_ZCU104/ \
--net_name otonixWe have setup PYNQ on our ZCU104, you can follow the guide here https://pynq.readthedocs.io/en/v2.6.1/getting_started/zcu104_setup.html,
The Juypter notebook launches automatically at the end of the platform boot. We access the Juypter notebook on a browser at http://board_ip_address. We gather the board_ip_address by accessing it via jtag-over-usb from a laptop.
Once the board is up and running we create the ear otoscopy application in the Juypter notebook environment. Vitis AI applications can be created using C++ or Python APIs. All required API libraries can be installed in the board image, follow here https://www.hackster.io/wadulisi/easy-ai-with-python-and-pynq-dd4822. We create the application using Python APIs. The first step of creating a Vitis AI application in Python is to transform the DPU object file, dpu_otonix_0.elf, we generated in the network compilation step above into a shared library. We achieve this by using the Arm GCC toolchain. Refer to the Juypter notebook otonix_app.ipnyb. The naming format of the shared library must be “libdpumodelModelName.so”, in our case its libdpumodelotonix.so, We place the shared library in the /dpuv2_rundir/ subdirectory which is in the same path level of the otonixl_app.ipynb script. The command used is as follows:
# Transform the dpu_otonix_0.elf into shard library libdpumodelotonix.so
!/usr/bin/aarch64-xilinx-linux-gcc -fPIC -shared ./coutput/output_ZCU104/dpu_otonix_0.elf -o libdpumodelotonix.soVitis AI comes with a ‘Runner Class’ which provides the abstraction for FPGA initialization and methods for scheduling compute tasks to the DPU.
# Instantiate the runner for the DPU
dpu = runner.Runner(path+'/dpuv2_rundir/' )we gather the input and output tensors of the DPU using the following API calls. The outputs are in Python list format.
# get the input and output tensors
inputTensors = dpu.get_input_tensors()
outputTensors = dpu.get_output_tensors()We estimate the input and output dimensions from the input and output Tensors lists as follows:
shapeIn = (batchSize,) + tuple([inputTensors[0].dims[i] for i in range(inputTensors[0].ndims)][1:])
outputHeight = outputTensors[0].dims[2]
outputWidth = outputTensors[0].dims[3]
outputChannel = outputTensors[0].dims[1]
We use these dimensions to create input and output data buffers in the memory:
outputData.append(np.empty((batchSize,outputSize),dtype=np.float32,order='C'))
inputData.append(np.empty((shapeIn), dtype = np.float32, order = 'C'))We then populate the input buffer with the otoscopic images we want to do detection as follows:
#place input images in to the input buffer
k = steps * batchSize
for j in range(batchSize):
imageRun = inputData[0]
imageRun[j,...]
=img[j+k].reshape(inputTensors[0].dims[1],inputTensors[0].dims[2],inputTensors[0].dims[3])Finally, we schedule the computing task to the DPU with the location of the input buffer and the output buffer, where it dumps the computed outputs. Once DPU execution completes the results are available in the outputData buffer.
dpu.execute_async(inputData,outputData)Finally, we execute the Softmax computation in the CPU by calling CPUCalcSoftmax(..) with the outputData. The CPUCalcSoftmax(..) is a Python function defined in the otonix_dataaug.py file.
CPUCalcSoftmax(outputData[0][j], outputSize)The output of the Softmax function is a list of detected classes of the batch of ear otoscopy images we fed via inputData buffer. See the code section for inference ipynb file.
Future Works:Make GUI for the model for quick inference load-up and as a commercial solution. Also, train the model on a heavy cloud or host system to achieve maximum accuracy. Further, we would also like to do segmentation on top of images to understand and classify all types of ear diseases with accuracy.
Thanks for reading this project.












Comments