THIS IS ONLY A PROTOTYPE, AND I AM NOT A MEDICAL PROFESSIONAL. PLEASE CONSULT A MEDICAL PROFESSIONAL BEFORE ATTEMPTING TO USE THIS DEVICE FOR MEDICATION ADMINISTRATION
Persons with visual impairments have to overcome many obstacles that arise from tasks that many find trivial. One such challenge is ensuring that proper medication is administered in the proper dosages. Pill form medications are small in size and difficult to read even with 20/20 vision. Modern advances in machine learning and micro processors give us the tools we need to build devices that can aid the visually impaired with these every day, important tasks.
PILBOIPILBOI PillIdentification, Logistics, Binning with Optical Inference is a home automation device capable of not only determining a medication type, but ensuring that the correct medication, in the correct dosage is determined. Using YOLO v8 object detection, PILBOI is able to discover pills in a field of view, maneuver the pill to an ideal inference location, infer the type of the medication and transfer the medication to one of two bins depending on the user's needs.
The device is programmable to enable a user to program the exact pill type and quantity. After the user specifies the desired pill collection, the information is uploaded and ready for use.
PILBOI in action:
StatesOnce the device is powered on, PILBOI enters its duty cycle. The cycle can be summarized as:
Initially the pill delivery stepper motor is incremented to deliver one pill to the imaging platform. Once the pill has been delivered, the imaging stepper motor is incremented in counter-clockwise steps until the Seeed Grove AI V2 MCU + PILBOI software has determined a pill is in the field of view. Next, the MCU will determine how to transition the pill to the ideal position for inference. Once the pill is in position, the PILBOI YOLO algorithm is run to determine the pill type. After the pill type is determined, PILBOI will transition the pill to the appropriate pill bin and continue on to the next pill in the delivery pipeline.
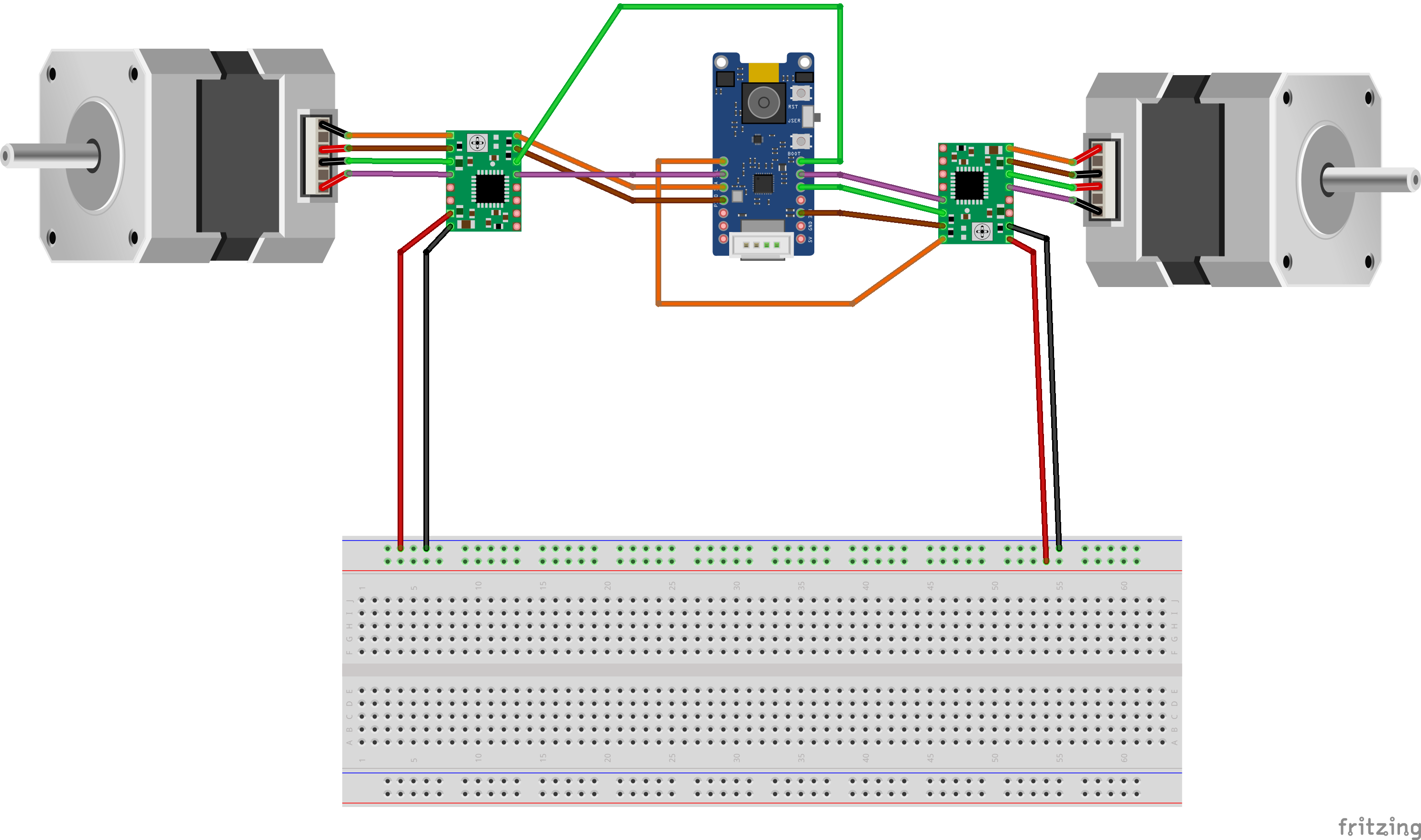
AssemblyThe following describes how to assemble PILBOI. Once the 3d parts are printed (see parts listing below), proceed to wiring the two stepper motors (see wiring diagram schematics below). Now you're ready to wire the stepper motor controllers to the appropriate Grove Vision AI2 GPIOs (again, please see wiring schematics). Follow the pictures below to install the stepper motors into the stepper motor chassis and mount the camera on the camera arm. Once motors and camera are mounted, proceed to installing the pill table and pill wheel to motor heads. Now you're ready to mount the pill wheel motor mount and the camera arm to the PILBOI body.
PILBOI YOLO V8 model in action as captured using Sensecraft.
The Sensecraft SSCMA framework was used to create the YOLO model used by PILBOI for pill object detection and identification. SSCMA provides many useful scripts and examples (to include Google Colab notebooks) that one can use to generate a YOLO V8 quantized model that utilizes the onboard Ethos NPU.
The PILBOI dataset consists of 9 classes, made up of pills of various sizes, colors and shapes. The 9 classes are:
1: Vitamin C
2: Lipitor
3: Vinegar
4: Acetaminophen
5: Airborne
6: Centrum
7: Iron
8: Magnesium
9: Pills (general)
The YOLO model accuracy is as follows:
Average Precision (AP) @[ IoU=0.50:0.95 | area= all | maxDets=100 ] = 0.642
Average Precision (AP) @[ IoU=0.50 | area= all | maxDets=100 ] = 0.923
Average Precision (AP) @[ IoU=0.75 | area= all | maxDets=100 ] = 0.823
Average Precision (AP) @[ IoU=0.50:0.95 | area= small | maxDets=100 ] = 0.418
Average Precision (AP) @[ IoU=0.50:0.95 | area=medium | maxDets=100 ] = 0.641
Average Precision (AP) @[ IoU=0.50:0.95 | area= large | maxDets=100 ] = 0.729
Average Recall (AR) @[ IoU=0.50:0.95 | area= all | maxDets= 1 ] = 0.691
Average Recall (AR) @[ IoU=0.50:0.95 | area= all | maxDets= 10 ] = 0.695
Average Recall (AR) @[ IoU=0.50:0.95 | area= all | maxDets=100 ] = 0.695
Average Recall (AR) @[ IoU=0.50:0.95 | area= small | maxDets=100 ] = 0.417
Average Recall (AR) @[ IoU=0.50:0.95 | area=medium | maxDets=100 ] = 0.693
Average Recall (AR) @[ IoU=0.50:0.95 | area= large | maxDets=100 ] = 0.764PILBOI dataset is freely available from roboflow. The dataset consists of 1531 images, 2063 annotations with the following class counts:
The PILBOI YOLO model is generated using Google Colab (runtime T4 GPU) with the SSCMA Jupyter Notebook downloadable from sensecraft. See code section for full Notebook. While the Seeed Grove Vision AI V2 is very capable device, it's limited on processing and memory (see Grove pages for details). As such, a standard YOLO model won't fit on the AI V2 device. Quantization and vela Ethos optimization must be applied to ensure the model is sized to fit on the AI V2 device. The provided Notebook from Sensecraft has the utilities to build the model in this fashion.
The steps to generate the PILBOI YOLO model are as follows.
- Download PILBOI dataset
- Load PILBOI Notebook into Google Colab
- Customize Notebook for epochs, dataset etc.
- Run the Notebook and download the resulting vela model.
The PILBOI software is a fork from HimaxWiseEyePlus Seeed Grove Vision AI Module V2 . PILBOI is freely available in GitHub.
There are two applications for PILBOI; pilboi-train and pilboi-yolo. pilboi-train is an application to generate training data for PILBOI and not covered in detail here. pilboi-yolo is the main PILBOI application and provides all of the features described in this writeup.
Pilboi-yolo has code to load the YOLO model, run two sets of stepper motors and execute the PILBOI algorithm to identify, classify and account pill based medication. The major software components are as follows:
Model Ingest
The model is loaded to a well known location in flash 0x00200000. At runtime, the software will load the model that is flashed to this location, and a TFLite model is created:
cvapp_yolov8nob.cpp
static const tflite::Model *yolov8n_ob_model = tflite::GetModel((const void *)model_addr);
if (yolov8n_ob_model->version() != TFLITE_SCHEMA_VERSION)
{
xprintf(
"[ERROR] yolov8n_ob_model's schema version %d is not equal "
"to supported version %d\n",
yolov8n_ob_model->version(), TFLITE_SCHEMA_VERSION);
return -1;
}
else
{
xprintf("yolov8n_ob model's schema version %d\n", yolov8n_ob_model->version));
}
static tflite::MicroErrorReporter yolov8n_ob_micro_error_reporter;
static tflite::MicroMutableOpResolver<13> yolov8n_ob_op_resolver;
yolov8n_ob_op_resolver.AddQuantize();
yolov8n_ob_op_resolver.AddGather();
yolov8n_ob_op_resolver.AddTranspose();
yolov8n_ob_op_resolver.AddConv2D();
yolov8n_ob_op_resolver.AddDepthwiseConv2D();
yolov8n_ob_op_resolver.AddAdd();
yolov8n_ob_op_resolver.AddRelu6();
yolov8n_ob_op_resolver.AddResizeNearestNeighbor();
yolov8n_ob_op_resolver.AddReshape();
yolov8n_ob_op_resolver.AddConcatenation();
yolov8n_ob_op_resolver.AddLogistic();
yolov8n_ob_op_resolver.AddPadV2();
if (kTfLiteOk != yolov8n_ob_op_resolver.AddEthosU())
{
xprintf("Failed to add Arm NPU support to op resolver.");
return false;
}PILBOIAlgorithm
The PILBOI algorithm (see States above) is implemented using the Himax event_handler system. The various PILBOI states are implemented as events and event handlers. The complete set of PILBOI events are:
EVT_INDEX_PILBOI_BTN_DOWN,
EVT_INDEX_PILBOI_BTN_UP,
EVT_PILBOI_NEXT,
EVT_PILBOI_SEARCH,
EVT_PILBOI_CENTERED,
EVT_PILBOI_OD_DONE,
EVT_PILBOI_PILL_NOT_FOV,
EVT_PILBOI_PILL_FOV,
EVT_PILBOI_PILL_NOT_ID,
EVT_PILBOI_PILL_ID,
EVT_PILBOI_PILL_CENTER,
EVT_PILBOI_BAD_PILL,
EVT_PILBOI_GOOD_PILL,As PILBOI transitions states, the appropriate event handlers are invoked. The complete set of events and callbacks can be found at the Github link below.
PILBOI Configuration
The PILBOI can be configured by running the pilboi-cfg.sh script. The script provides the desired pill counts for all pills of interest.
#!/bin/bash
#C
echo -n -e \\x00\\x00 > ~/pilboi-cfg.bin
#lip
echo -n -e \\x01\\x00 >> ~/pilboi-cfg.bin
#vin
echo -n -e \\x00\\x00 >> ~/pilboi-cfg.bin
#ace
echo -n -e \\x00\\x00 >> ~/pilboi-cfg.bin
#air
echo -n -e \\x00\\x00 >> ~/pilboi-cfg.bin
#cen
echo -n -e \\x01\\x00 >> ~/pilboi-cfg.bin
#iron
echo -n -e \\x01\\x00 >> ~/pilboi-cfg.bin
#mag
echo -n -e \\x02\\x00 >> ~/pilboi-cfg.bin
#dd < /dev/zero bs=4080 count=1 >> ~/pilboi-cfg.binAfter running the script, a binary file (in big endian) will be produced pilboi-cfg.bin. This file needs to be flashed to 0x00300000 and can be done so using the xmodem_send.py script provided.
python3 xmodem/xmodem_send.py --port=/dev/ttyACM0 --baudrate=921600 --protocol=xmodem --file=we2_image_gen_local/output_case1_sec_wlcsp/output.img --model="/home/foo/models/epoch_100_combine_int8_vela.tflite 0x00200000 0x0000" --model="/home/foo/pilboi-cfg.bin 0x00300000 0x0000"PILBOIMath
Determining the pill locations and motor positions requires a bit of math. The problem we are trying to solve is centering a pill in the camera's field of view. We start by defining 3 points, the camera center, the pivot center, and the pill centroid. From these points we then find the distance using the distance formula:
To find out how far we have to rotate the pill we use the law of cos to determine an angle in degrees:
Once the desired angle is determined, PILBOI will command the pill imaging stepper motor to desired location.
All the PILBOI parts can be printed using a standard 3d printer (Ender 3 pro in my case) and are freely available below. The PILBOI parts are:
Pill wheel motor mount - Houses the pill wheel motor
Pill bed motor mount - Houses the pill bed motor
Pill wheel - Used to transfer pill from pill feeder to pill bed
Pill bed - Used to image and maneuver pill to be ID'd
Camera arm - Used to mount the camera
Pill tray - Used to catch and separate both bad and good pills
PILBOI chassis - The main PILBOI chassis
ConclusionThank you so much for your attention to this point. Working on PILBOI was challenging and rewarding. While I see huge potential in helping to identify, account for and administer pill based medication with the aid of AI, there's still a ways to go. The test case proved there is huge potential, but give the vast number pill shapes and sizes, much more data is needed to achieve a production worthy dataset.





















_3u05Tpwasz.png?auto=compress%2Cformat&w=40&h=40&fit=fillmax&bg=fff&dpr=2)
Comments