Interactions between cells in complex communities can result in the emergence of unexpected functional behaviors beyond what a simple cellular monoculture could achieve. There is substantial untapped potential in exploiting cellular communities to advance a range of synthetic biology applications, from biomaterials to more efficient cellular factories. We are building a toolkit for systematically engineering the structural and ecological interactions that can occur between mixed populations of two cell types. Our toolkit will provide both artificial surface-receptors for building combinations of adhesive interactions between Escherichia coli and modular circuits for generating synergistic and antagonistic ecological relationships between cells. With this set of intercompatible and functionally-characterized parts, we will provide an open and accessible resource for scientists, educators, and community-based labs to begin programming multi-cellular bacterial communities.
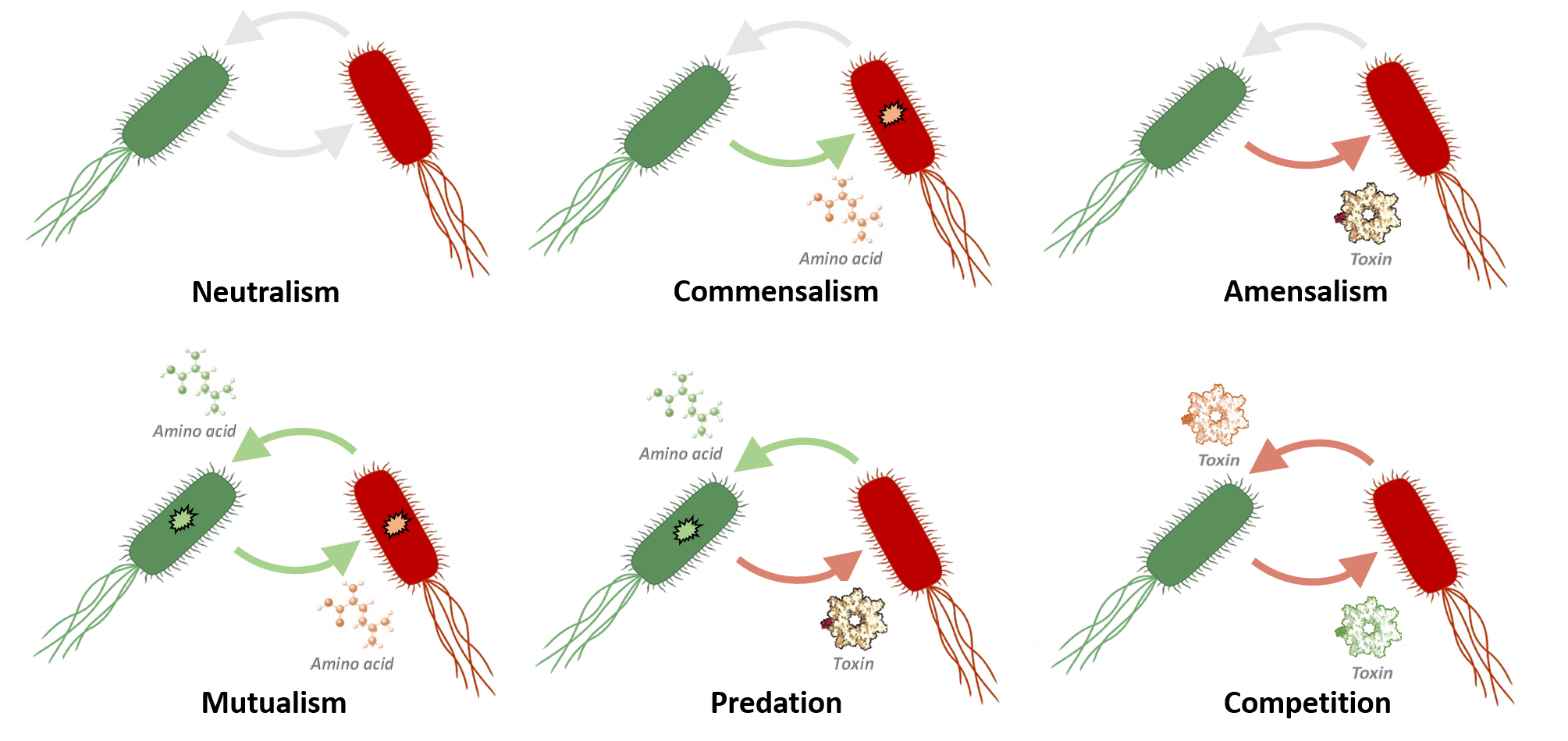
Biological building-blocksTo engineer bacterial systems with defined interactions, we take advantage of 3 main biological "building-blocks". Those are 1) synthetic genes encoding proteins that allow cells to selectively recognize and bind other cells (termed "nanobodies"), 2) genetically-mutated strains of bacteria that cannot produce certain essential nutrients on their own (known as "auxotrophs"), and 3) portable DNA elements known as plasmids that generate antibacterial toxins and selective immunity to those toxins.
Nanobody adhesion receptors
We have several bacterial strains transformed with plasmids which code for nanobody-antigen pairs. These nanobody proteins are expressed on the surface of bacterial cells which have been successfully transformed with the appropriate plasmid. Other bacterial cells will display antigens on their cell surface if they have been transformed with the appropriate plasmid.
Nanobodies will bind with high specificity and affinity to a particular surface antigen, and so different cells displaying complementary nanobody-antigen pairs will stick to one another and form clumps. These cells are also tagged with different fluorescent proteins so that emergent structure can be detected analysed under the microscope.
Auxotrophic mutants
We have 8 E. coli mutants which are auxotrophic for 4 different amino acids (tryptophan, methionine, proline and isoleucine) and are tagged with either eGFP or mCherry.
Each auxotroph will be unable to grow unless the media it is in is supplemented with the relevant amino acid, and the growth of these auxotrophs can be detected by their fluorescence (either in a plate reader of under a microscope), or by observing turbidity (either in a plate reader or spectrophotometer).
Amino acids will diffuse out E. coli cells, and so different auxotrophs in the same environment may be able to share resources with one another, sustaining growth in media with no supplemented amino acid. This can be used to generate mutualistic interaction pairs.
Toxin-antitoxin systems
Bacteria have evolved natural systems by which they can secrete toxins, which are large molecules used to kill other cells. Bacteria can be transformed with plasmids which code for the production of a toxin-antitoxin pair, thus conferring resistance to the toxin to the toxin-producing bacteria. These toxins which are released into the surrounding media will harm other bacteria without the antitoxin, thus producing antagonistic social interactions between cells.
Results (as of 2019.10.30)Setting up growth conditions for mutant strains
We first had to tune growth conditions so the auxotrophic mutant E.coli in our toolkit will be unable to grow on their own unless media is supplemented. After failing to see any growth on a strict media formulation with limited carbon sources, we saw reasonable growth with M9 minimal media supplemented with 0.4% glucose. We could rescue growth of our methionine, isoleucine, proline, and tryptophan auxotrophs by supplementing each with an excess (i.e. 100 ug/mL) of their respective amino acids (Fig 1A). On supplemented and unsupplemented plates, we confirmed that other amino acids could not be substituted for any of our auxotrophs, and that the growth observed was not merely from contamination (Fig 1B). To observe these effects, we had to inoculate relatively small amounts of bacteria (<3 uL). Higher inoculation amounts lead to nonspecific growth of mutants on all media conditions, likely because of amino acids released by dead cells.
Establishing symbiotic cultures of auxotrophs
With the functional identities of our auxotrophic strains verified, we next moved on to mixing pairs of strains together to identify if strains that are otherwise non-viable can, when mixed, restore each others' ability to grow through a mutualistic interaction. We found that these experiments are very sensitive to starting cell numbers, carryover of growth media into the minimal media co-culture chamber, and differences in each auxotrophs' viability. However, we were able to create strong mutualistic interactions between certain auxotroph pairs, including growth of -Methionine & -Isoleucine pairs and no growth of auxotrophs of the same background such as -Isoleucine & -Isoleucine (Fig 2). We are currently testing other conditions for setting up our co-cultures to correct for viability failures in our first experiments (such as the red fluorescence-tagged -Proline strain) and unexpected growth in conditions where auxotrophs should die (such as the tryptophan-mutant strains, not shown).
Measuring genome-encoded fluorescence tags
To distinguish the strains we are co-culturing, we selected compatible strains with different fluorescent protein markers integrated in their genomes (mCherry for red fluorescence, and eGFP for green fluorescence). We put slides of these strains and negative control (non-fluorescent) and positive-control (known fluorescent) bacteria at different densities under an epifluorescence microscope. We found a dilution density that results in dispersed single-cells (as opposed to clumps or stacking) and plenty of cells in the field of view (as opposed to sparse views with too few cells to analyze). After calibrating the microscope settings on the positive controls, we saw clear fluorescent signals on all of our eGFP and mCherry-labeled strains. However, we also experienced extremely rapid photobleaching, with signals vanishing to undetectable levels within seconds of illumination. Although none of several anti-fade mounting mixtures we tested could eliminate this problem, we were able to extend the fluorescence lifetime by several seconds, enough to allow careful imaging of cells (Fig 3).
Validating nanobody adhesive strains
We established a protocol for introducing our adhesion plasmids into our many different tagged and auxotrophic E. coli backgrounds without the usual first step of bacterial transformation where cells are first made competent. Our electroporation-based protocol saves considerable time in making genetically-modified bacteria out of numerous strains, and has the added benefit of avoiding equipment like refrigerated centrifuges and access to flash-freezing that are not accessible to many community lab spaces like ours.
Using this protocol, we have generated strains harboring adhesion plasmids and fluorescent tags, plus a small selection of auxotrophs with adhesion plasmids. Before transforming, we performed Sanger sequencing on all the adhesion plasmids to verify each construct exactly matches the intended sequences. This is particularly important given some of the peptides are only 4 amino acids long, and thus would be otherwise difficult to detect errors in.
We have begun testing the set of surface-displayed nanobody and antigen pairs we have for programming physical interactions. Through a simple clumping assay, we can observe slight changes in the turbidity of cultures where bacteria are mixed with binding nanbody-antigen sets, due to the binding cells aggregating into dense clumps that can sediment. We have seen encouraging preliminary results for one out of our three adhesion pairs (Fig 4A). In two our of three replicates of a mixture of nanobody-expressing cells and paired antigen-expressing cells there was dramatic clumping within the first hour, while no controls without a nanobody match had significant binding (Fig 4B).
Antagonistic interactions with a toxin/antitoxin system
We have also designed a plasmid for generating bacteria that secrete toxins to which they themselves are immune to by the internal expression of an antitoxin (Fig 5). We selected colicin E1 for this toxin-antitoxin plasmid, and under the regulation of an inducible arabinose promoter to be able to titrate toxin levels so that interesting ecological interactions can take place, such as amensalism, without being so strong that cells without the antitoxin cannot grow and interact. We have so far extracted out half of the parts needed for our final plasmid assembly, and will clone and test the plasmid using our established growth assays.
Co-culture phenotypes under the microscope
With a large fraction of our base strains prepared and validated, we proceeded to characterize how co-cultures of interacting cells appear at the microscopic level. As a first example, we took mutually interdependent auxotrophic mutants and let them establish stable growth together. One mutant we labeled in green (eGFP) and the other in red (mCherry). After transferring the cells from our culture plates onto microscopy slides, we could view the distinct cell populations under a widefield fluorescence microscope (Fig 6A). There are a large number of phenotypes that could be extracted from images of our co-cultures, such as cell proximity, ratios, and localization (Fig 6B). Although we have not yet begun to fully explore these potential analyses of our imaging data, we intend to work on methods to quantify co-culture images.
How to use our microbial toolkit
For end users interested in trying our toolkit, we provide here an example illustrating how the toolkit works and the corresponding protocols and resources we’ve made available. In its final form, users will be sent aliquots of frozen glycerol stocks we have prepared for each E. coli strain in the toolkit. The strains include bacteria with the different combinations of auxotrophic mutations, inducible toxin/antitoxin plasmids, and surface-displayed nanobodies and antigens. Each strain has either a green or red fluorescent tag in its genome for easy identification in experiments.
By mixing two strains together, users can set up a defined two-celltype community. Our protocols cover all the basic necessary information needed to grow up the bacterial stocks into cultures, and ways to mix strains on microtiter plates for plate-based experiments. Following these protocols, we have reliably been able to get proper growth behavior even for the auxotrophic mutants. Importantly, these guidelines allow mutualistic and commensal interactions to be observed.
Protocols are also provided for inducing cell aggregation with our surface adhesion proteins. Once our toxin/antitoxin system is finalized, we will also have instructions for calibrating the level of induction to get stable antagonistic interactions. We also have a procedure for monitoring the growth of bacteria within co-cultures using their fluorescent tags, which is particularly useful for ecological interactions. Python code for analyzing growth data is provided alongside our protocols.
Any of these protocols can be used to individually characterize a given co-culture. We also have protocols for preparing samples of cells for visualizing under a microscope, which we expect will be a particularly interesting source of observations for users to explore at increasing levels of microbial community complexity.
OutlookWe hope to soon finish assembling the adhesive and mutant strains that will comprise our interaction toolbox. The protocols we are working to optimize will be another key component of this toolkit, so others can easily establish the proper culturing conditions to observe interesting biological effects.
Our toolkit has many potential applications. First, it may find use as an educational resource, to show microbial interactions in simple settings with easy to view readouts. Second, they could be research tools for looking at microbial interactions in a reductionist framework. Possibilities of complex cell self-assembly are particularly interesting, and have data from others supporting its plausibility. Third, having stable interactions between mixed cell populations could be used to optimize bioreactors beyond what can be reached with a monoculture. Lastly, being able to program these interactions could turn cells into scaffolds for producing patterned biomaterials. Through both our work as part of the Biomaker Challenge and our projects with students as the Cambridge University Synthetic Biology Society, we hope to continue developing these tools towards these exciting applications.
ReferencesGlass DS, Riedel-kruse IH. A Synthetic Bacterial Cell-Cell Adhesion Toolbox for Programming Multicellular Morphologies and Patterns. Cell. 2018;174(3):649-658.e16.
Jakes KS. The Colicin E1 TolC Box: Identification of a Domain Required for Colicin E1 Cytotoxicity and TolC Binding. J Bacteriol. 2017;199(1)
Marchal M, Goldschmidt F, Derksen-müller SN, Panke S, Ackermann M, Johnson DR. A passive mutualistic interaction promotes the evolution of spatial structure within microbial populations. BMC Evol Biol. 2017;17(1):106.
Kong W, Meldgin DR, Collins JJ, Lu T. Designing microbial consortia with defined social interactions. Nat Chem Biol. 2018;14(8):821-829.







Comments
Please log in or sign up to comment.