The Acute Lymphoblastic Leukemia Arduino Nano 33 BLE Sense Classifier is an experiment to explore how low powered microcontrollers, specifically the Arduino Nano 33 BLE Sense, can be used to detect Acute Lymphoblastic Leukemia. The Arduino Nano 33 BLE Sense is the latest Arduino Board which supports Tensorflow Lite, allowing machine learning on Arduino.
The model you will train is a 6 layer Convoluntional Neural Network trained using Intel® Optimization for Tensorflow* from the Intel® oneAPI AI Analytics Toolkit to optimize and accelerate the training process.
DISCLAIMERThis project should be used for research purposes only. The purpose of the project is to show the potential of Artificial Intelligence for medical support systems such as diagnostic systems.
Although the model is accurate and shows good results both on paper and in real world testing, it is trained on a small amount of data and needs to be trained on larger datasets to really evaluate it's accuracy.
Developers that have contributed to this repository have experience in using Artificial Intelligence for detecting certain types of cancer. They are not doctors, medical or cancer experts.
MotivationThe motivation for this project was to explore how low powered devices such as Arduino can be used to detect Acute Lymphoblastic Leukemia. The project will be submitted to the Tensorflow For Microcontroller Challenge and the Eyes on Edge: tinyML Vision Challenge.
Acute Lymphoblastic LeukemiaAcute lymphoblastic leukemia (ALL), also known as Acute Lymphocytic Leukemia, is a cancer that affects the lymphoid blood cell lineage. It is the most common leukemia in children, and it accounts for 10-20% of acute leukemias in adults. The prognosis for both adult and especially childhood ALL has improved substantially since the 1970s. The 5- year survival is approximately 95% in children. In adults, the 5-year survival varies between 25% and 75%, with more favorable results in younger than in older patients.
For more information about Acute Lymphoblastic Leukemia please visit our Leukemia Information Page
ALL-IDBYou need to be granted access to use the Acute Lymphoblastic Leukemia Image Database for Image Processing dataset. You can find the application form and information about getting access to the dataset on this page as well as information on how to contribute back to the project here. If you are not able to obtain a copy of the dataset please feel free to try this tutorial on your own dataset, we would be very happy to find additional AML & ALL datasets.
Clone the repositoryClone the ALL Arduino Nano 33 BLE Sense Classifier repository from the Peter Moss Acute Myeloid & Lymphoblastic Leukemia AI Research Project Github Organization.
To clone the repository and install the project, make sure you have Git installed. Now navigate to the directory you would like to clone the project to and then use the following command.
git clone https://github.com/AMLResearchProject/ALL-Arduino-Nano-33-BLE-Sense-Classifier.gitThis will clone the ALL Arduino Nano 33 BLE Sense Classifier repository.
lsUsing the ls command in your home directory should show you the following.
ALL-Arduino-Nano-33-BLE-Sense-ClassifierNavigate to the ALL-Arduino-Nano-33-BLE-Sense-Classifier directory, this is your project root directory for this tutorial.
Developer forksDevelopers from the Github community that would like to contribute to the development of this project should first create a fork, and clone that repository. For detailed information please view the CONTRIBUTING guide. You should pull the latest code from the development branch.
git clone -b "2.0.0" https://github.com/AMLResearchProject/ALL-Arduino-Nano-33-BLE-Sense-Classifier.gitThe -b "2.0.0" parameter ensures you get the code from the latest master branch. Before using the below command please check our latest master branch in the button at the top of the project README.
InstallationYou are now ready to install the ALL Arduino Nano 33 BLE Sense Classifier trainer. All software requirements are included in scripts/install.sh. You can run this file on your machine from the project root in terminal. Use the following command:
sh scripts/install.shWARNING: This script assumes you have not already installed the oneAPI Basekit.
WARNING: This script assumes you have not already installed the oneAPI AI Analytics Toolkit.
WARNING: This script assumes you have an Intel GPU.
WARNING: This script assumes you have already installed the Intel GPU drivers.
HINT: If any of the above are not relevant to you, please comment out the relevant sections in the installation script before running it.
DataYou need to be granted access to use the Acute Lymphoblastic Leukemia Image Database for Image Processing dataset. You can find the application form and information about getting access to the dataset on this page as well as information on how to contribute back to the project here.
If you are not able to obtain a copy of the dataset please feel free to try this tutorial on your own dataset.
Once you have your data you need to add it to the project filesystem. You will notice the data folder in the Model directory, model/data, inside you have train & test. Add all of the images from the ALL_IDB1 dataset to the model/data/train folder.
Data AugmentationWe will create an augmented dataset based on the Leukemia Blood Cell Image Classification Using Convolutional Neural Network by T. T. P. Thanh, Caleb Vununu, Sukhrob Atoev, Suk-Hwan Lee, and Ki-Ryong Kwon.
Application testing dataIn the data processing stage, ten negative images and ten positive images are removed from the dataset and moved to the model/data/test/ directory. This data is not seen by the network during the training process, and is used to test the performance of the model.
To ensure your model gets the same results, you should use the same test images. You can also try with your own image selection, however results may vary.
To specify which test images to use modify the configuration/config.json file as shown below:
"test_data": [
"Im006_1.jpg",
"Im020_1.jpg",
"Im024_1.jpg",
"Im026_1.jpg",
"Im028_1.jpg",
"Im031_1.jpg",
"Im035_0.jpg",
"Im041_0.jpg",
"Im047_0.jpg",
"Im053_1.jpg",
"Im057_1.jpg",
"Im060_1.jpg",
"Im063_1.jpg",
"Im069_0.jpg",
"Im074_0.jpg",
"Im088_0.jpg",
"Im095_0.jpg",
"Im099_0.jpg",
"Im101_0.jpg",
"Im106_0.jpg"
],All configuration can be found in the configuration/config.json file.
{
"agent": {
"cores": 8,
"ip": "",
"port": 1234,
"params": [
"train",
"classify",
"server",
"classify_http"
]
},
"data": {
"dim": 100,
"file_type": ".jpg",
"labels": [0, 1],
"rotations": 10,
"seed": 2,
"split": 0.255,
"test": "model/data/test",
"test_data": [
"Im006_1.jpg",
"Im020_1.jpg",
"Im024_1.jpg",
"Im026_1.jpg",
"Im028_1.jpg",
"Im031_1.jpg",
"Im035_0.jpg",
"Im041_0.jpg",
"Im047_0.jpg",
"Im053_1.jpg",
"Im057_1.jpg",
"Im060_1.jpg",
"Im063_1.jpg",
"Im069_0.jpg",
"Im074_0.jpg",
"Im088_0.jpg",
"Im095_0.jpg",
"Im099_0.jpg",
"Im101_0.jpg",
"Im106_0.jpg"
],
"train_dir": "model/data/train",
"valid_types": [
".jpg"
]
},
"model": {
"model": "model/all_nano_33_ble_sense.json",
"model_c_array": "model/all_nano_33_ble_sense.cc",
"tfmodel": "model/all_nano_33_ble_sense.tflite",
"weights": "model/all_nano_33_ble_sense.h5"
},
"train": {
"batch": 100,
"decay_adam": 1e-6,
"epochs": 150,
"learning_rate_adam": 1e-4,
"val_steps": 10
}
}You should update the following values:
- agent->cores Should represent the amount of cores your CPU has.
- agent->ip Should be the IP of the machine you will run your test server on.
- agent->port Should be the port you will run your test server on.
You can modify the values in the train object as required, however to ensure you achieve the same results you can leave them as they are.
TrainingNow you are ready to train your model.
MetricsWe can use metrics to measure the effectiveness of our model. In this network you will use the following metrics:
tf.keras.metrics.BinaryAccuracy(name='accuracy'),
tf.keras.metrics.Precision(name='precision'),
tf.keras.metrics.Recall(name='recall'),
tf.keras.metrics.AUC(name='auc')These metrics will be displayed and plotted once our model is trained. A useful tutorial while working on the metrics was the Classification on imbalanced data tutorial on Tensorflow's website.
Start TrainingEnsuring you have completed all previous steps, you can start training using the following command.
python classifier.py trainThis tells the application to start training the model. You can also use the Jupyter Notebook to train your model. If you choose to do so you can access the Notebook here. Checkout the documentation here.
Training DataFirst the training and validation data will be prepared.
2021-07-18 17:42:26,075 - Classifier - INFO - Augmented data size: 1584
2021-07-18 17:42:26,075 - Classifier - INFO - Negative data size: 882
2021-07-18 17:42:26,076 - Classifier - INFO - Positive data size: 702
2021-07-18 17:42:26,076 - Classifier - INFO - Augmented data shape: (1584, 100, 100, 3)
2021-07-18 17:42:26,076 - Classifier - INFO - Labels shape: (1584, 2)
2021-07-18 17:42:26,267 - Classifier - INFO - Training data: (1180, 100, 100, 3)
2021-07-18 17:42:26,267 - Classifier - INFO - Training labels: (1180, 2)
2021-07-18 17:42:26,267 - Classifier - INFO - Validation data: (404, 100, 100, 3)
2021-07-18 17:42:26,267 - Classifier - INFO - Validation labels: (404, 2)
2021-07-18 17:42:26,267 - Classifier - INFO - Data preperation complete.Before the model begins training, you will be shown the model summary.
Model: "AllANBS"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
average_pooling2d (AveragePo (None, 50, 50, 3) 0
_________________________________________________________________
conv2d (Conv2D) (None, 46, 46, 30) 2280
_________________________________________________________________
depthwise_conv2d (DepthwiseC (None, 17, 17, 30) 27030
_________________________________________________________________
flatten (Flatten) (None, 8670) 0
_________________________________________________________________
dense (Dense) (None, 2) 17342
_________________________________________________________________
activation (Activation) (None, 2) 0
=================================================================
Total params: 46,652
Trainable params: 46,652
Non-trainable params: 0
_________________________________________________________________
2021-07-18 17:42:26,323 - Classifier - INFO - Network initialization complete.
2021-07-18 17:42:26,324 - Classifier - INFO - Using Adam Optimizer.
2021-07-18 17:42:26,324 - Classifier - INFO - Using Early Stopping.Below are the training results for 28 epochs.
Fig 2. Accuracy
Fig 3. Loss
Fig 4. Precision
Fig 5. Recall
Fig 6. AUC
Fig 7. Confusion Matrix
2021-07-18 17:47:55,953 - Classifier - INFO - Metrics: loss 0.2371470034122467
2021-07-18 17:47:55,953 - Classifier - INFO - Metrics: acc 0.9331682920455933
2021-07-18 17:47:55,953 - Classifier - INFO - Metrics: precision 0.9331682920455933
2021-07-18 17:47:55,953 - Classifier - INFO - Metrics: recall 0.9331682920455933
2021-07-18 17:47:55,953 - Classifier - INFO - Metrics: auc 0.9677298069000244
2021-07-18 17:47:56,536 - Classifier - INFO - Confusion Matrix: [[217 4] [ 23 160]]
2021-07-18 17:47:56,633 - Classifier - INFO - True Positives: 160(39.603960396039604%)
2021-07-18 17:47:56,633 - Classifier - INFO - False Positives: 4(0.9900990099009901%)
2021-07-18 17:47:56,633 - Classifier - INFO - True Negatives: 217(53.71287128712871%)
2021-07-18 17:47:56,633 - Classifier - INFO - False Negatives: 23(5.693069306930693%)
2021-07-18 17:47:56,633 - Classifier - INFO - Specificity: 0.9819004524886877
2021-07-18 17:47:56,633 - Classifier - INFO - Misclassification: 27(6.683168316831683%)Accuracy 0.9331682920455933
Recall 0.9331682920455933
Precision 0.9331682920455933
AUC/ROC 0.9677298069000244
Figures Of MeritTrue Positives 160 (39.603960396039604%)
False Positives 4 (0.9900990099009901%)
True Negatives 217 (53.71287128712871%)
False Negatives 23 (5.693069306930693%)
Sensitivity / Recall 0.9331682920455933 (93%)
Specificity 0.9819004524886877 (98%)
TestingNow you will test the classifier on your training machine. You will use the 20 images that were removed from the training data in a previous part of this tutorial.
To run the classifier in test mode use the following command:
python3 classifier.py classifyYou should see the following which shows you the network architecture:
Model: "AllANBS"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
average_pooling2d (AveragePo (None, 50, 50, 3) 0
_________________________________________________________________
conv2d (Conv2D) (None, 46, 46, 30) 2280
_________________________________________________________________
depthwise_conv2d (DepthwiseC (None, 17, 17, 30) 27030
_________________________________________________________________
flatten (Flatten) (None, 8670) 0
_________________________________________________________________
dense (Dense) (None, 2) 17342
_________________________________________________________________
activation (Activation) (None, 2) 0
=================================================================
Total params: 46,652
Trainable params: 46,652
Non-trainable params: 0
_________________________________________________________________Finally the application will start processing the test images and the results will be displayed in the console.
2021-07-18 17:51:28,684 - Classifier - INFO - Loaded test image model/data/test/Im063_1.jpg
2021-07-18 17:51:28,804 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly detected (True Positive) in 0.12069535255432129 seconds.
2021-07-18 17:51:28,804 - Classifier - INFO - Loaded test image model/data/test/Im028_1.jpg
2021-07-18 17:51:28,838 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly detected (True Positive) in 0.03429007530212402 seconds.
2021-07-18 17:51:28,839 - Classifier - INFO - Loaded test image model/data/test/Im106_0.jpg
2021-07-18 17:51:28,872 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly not detected (True Negative) in 0.03346753120422363 seconds.
2021-07-18 17:51:28,872 - Classifier - INFO - Loaded test image model/data/test/Im101_0.jpg
2021-07-18 17:51:28,906 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly not detected (True Negative) in 0.034415245056152344 seconds.
2021-07-18 17:51:28,907 - Classifier - INFO - Loaded test image model/data/test/Im024_1.jpg
2021-07-18 17:51:28,939 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly detected (True Positive) in 0.032686471939086914 seconds.
2021-07-18 17:51:28,940 - Classifier - INFO - Loaded test image model/data/test/Im074_0.jpg
2021-07-18 17:51:28,972 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly not detected (True Negative) in 0.03266596794128418 seconds.
2021-07-18 17:51:28,973 - Classifier - INFO - Loaded test image model/data/test/Im035_0.jpg
2021-07-18 17:51:29,005 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly not detected (True Negative) in 0.032935142517089844 seconds.
2021-07-18 17:51:29,006 - Classifier - INFO - Loaded test image model/data/test/Im006_1.jpg
2021-07-18 17:51:29,039 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly detected (True Positive) in 0.03386235237121582 seconds.
2021-07-18 17:51:29,040 - Classifier - INFO - Loaded test image model/data/test/Im020_1.jpg
2021-07-18 17:51:29,076 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly detected (True Positive) in 0.0370943546295166 seconds.
2021-07-18 17:51:29,077 - Classifier - INFO - Loaded test image model/data/test/Im095_0.jpg
2021-07-18 17:51:29,109 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly not detected (True Negative) in 0.03277897834777832 seconds.
2021-07-18 17:51:29,110 - Classifier - INFO - Loaded test image model/data/test/Im069_0.jpg
2021-07-18 17:51:29,143 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly not detected (True Negative) in 0.03318381309509277 seconds.
2021-07-18 17:51:29,143 - Classifier - INFO - Loaded test image model/data/test/Im031_1.jpg
2021-07-18 17:51:29,175 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly detected (True Positive) in 0.03194856643676758 seconds.
2021-07-18 17:51:29,176 - Classifier - INFO - Loaded test image model/data/test/Im099_0.jpg
2021-07-18 17:51:29,208 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly not detected (True Negative) in 0.032364845275878906 seconds.
2021-07-18 17:51:29,208 - Classifier - INFO - Loaded test image model/data/test/Im026_1.jpg
2021-07-18 17:51:29,241 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly detected (True Positive) in 0.03291964530944824 seconds.
2021-07-18 17:51:29,241 - Classifier - INFO - Loaded test image model/data/test/Im057_1.jpg
2021-07-18 17:51:29,276 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly detected (True Positive) in 0.035039663314819336 seconds.
2021-07-18 17:51:29,277 - Classifier - INFO - Loaded test image model/data/test/Im088_0.jpg
2021-07-18 17:51:29,312 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly not detected (True Negative) in 0.03563833236694336 seconds.
2021-07-18 17:51:29,313 - Classifier - INFO - Loaded test image model/data/test/Im060_1.jpg
2021-07-18 17:51:29,346 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly detected (True Positive) in 0.03394198417663574 seconds.
2021-07-18 17:51:29,347 - Classifier - INFO - Loaded test image model/data/test/Im053_1.jpg
2021-07-18 17:51:29,383 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly detected (True Positive) in 0.03695321083068848 seconds.
2021-07-18 17:51:29,384 - Classifier - INFO - Loaded test image model/data/test/Im041_0.jpg
2021-07-18 17:51:29,417 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly not detected (True Negative) in 0.03377509117126465 seconds.
2021-07-18 17:51:29,418 - Classifier - INFO - Loaded test image model/data/test/Im047_0.jpg
2021-07-18 17:51:29,450 - Classifier - INFO - Acute Lymphoblastic Leukemia correctly not detected (True Negative) in 0.03243684768676758 seconds.
2021-07-18 17:51:29,450 - Classifier - INFO - Images Classified: 20
2021-07-18 17:51:29,450 - Classifier - INFO - True Positives: 10
2021-07-18 17:51:29,450 - Classifier - INFO - False Positives: 0
2021-07-18 17:51:29,450 - Classifier - INFO - True Negatives: 10
2021-07-18 17:51:29,450 - Classifier - INFO - False Negatives: 0
2021-07-18 17:51:29,450 - Classifier - INFO - Total Time Taken: 0.7630934715270996Here we trained a deep learning model for Acute Lymphoblastic Leukemia detection utilizing Intel® Optimization for Tensorflow* from the Intel® oneAPI AI Analytics Toolkit to optimize and accelerate training. We introduced a 6 layer deep learning model and applied data augmentation to increase the training data.
We trained our model with a target of 150 epochs and used early stopping to avoid overfitting. The model trained for 29 epochs resulting in a slightly noisy fit, however accuracy/precision/recall and AUC are satisfying. In addition the model reacts well during testing classifying each of the twenty unseen test images correctly.
ArduinoYou will need to ensure you have installed the following software and followed the provided guides:
- Ubuntu Installation Guide
- Python Usage Guide or Jupyter Notebooks Usage Guide
- Arduino IDE
- Getting started with the Arduino Nano 33 BLE Sense
- Why doesn't the 5V pin work in the Arduino Nano 33 BLE boards?
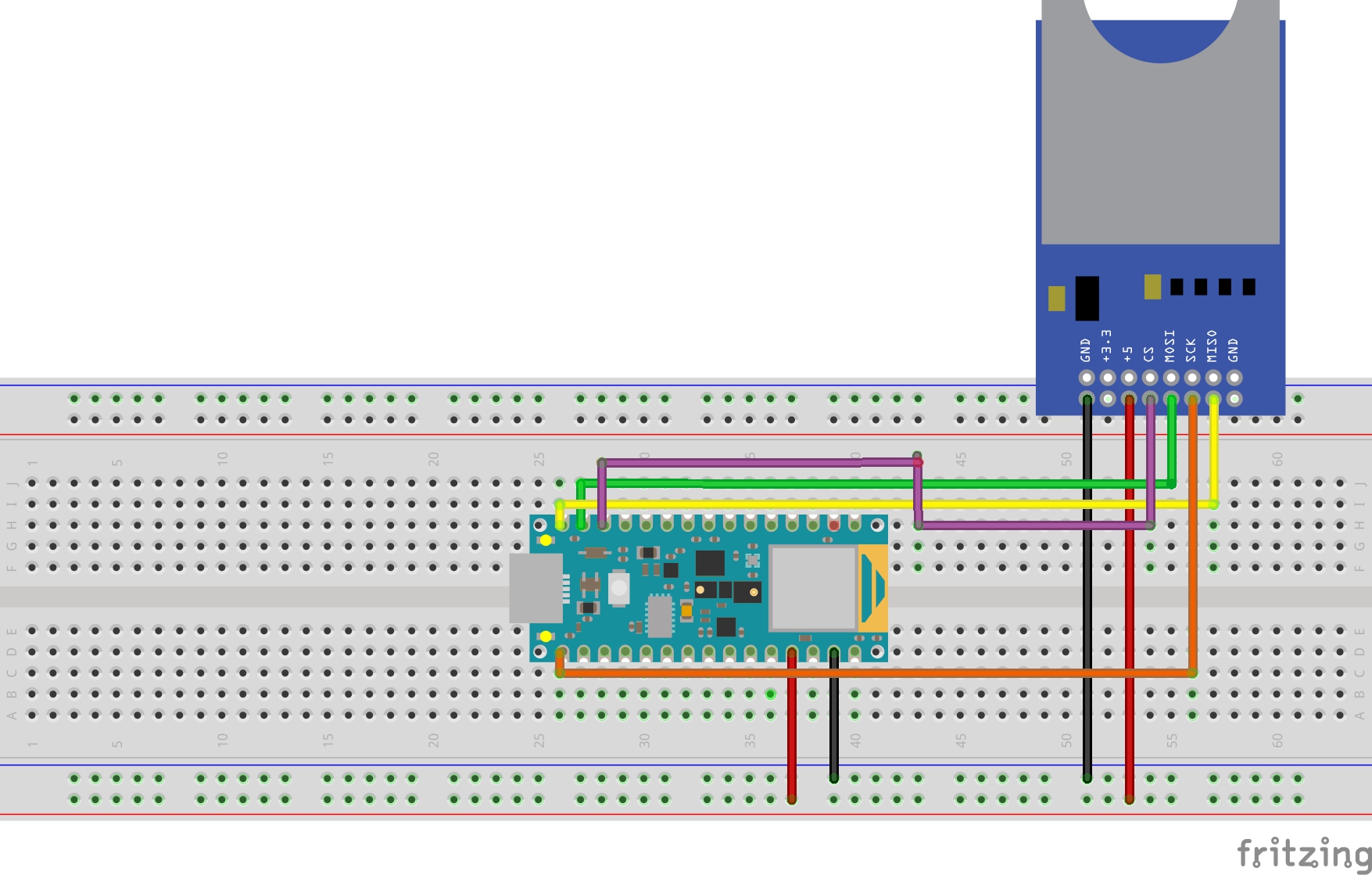
Follow the diagram above to connect your SD card reader to the Arduino Nano 33 BLE Sense.
Open your Arduino IDE and open the all_nano_33_ble_sense sketch located in the Arduino folder in the project root.
C Array ModelNow you need to import your C array model into the Arduino project. On your development machine navigate to the model dir located in the project root and open the all_nano_33_ble_sense.cc file. First you need to copy the model and replace everything within all_model[]{} with your newly created model. Next you need to replace all_model_len with the actual length of your model which is found at the bottom of your model file.
Test DataDuring training the test data was resized and moved to the **model/data/test/** directory. Before you can continue you need to upload these files to the SD card.
Run The ClassifierNow it is time to run your classifier on the Arduino Nano 33 BLE Sense. Make sure you are connected to your Arduino and click on the upload button. Once the model is uploaded it will start to run, open your serial monitor and watch the output.
19:22:40.139 -> Initialising SD card...
19:22:40.148 -> Initialisation done.
19:22:40.158 ->
19:22:40.163 -> Model input info
19:22:40.274 -> ===============
19:22:40.284 -> Dimensions: 4
19:22:40.299 -> Dim 1 size: 1
19:22:40.314 -> Dim 2 size: 100
19:22:40.328 -> Dim 3 size: 100
19:22:40.343 -> Dim 4 size: 3
19:22:40.354 -> Input type: 9
19:22:40.365 -> ===============
19:22:40.375 ->
19:22:40.381 -> Im006_1.jpg
19:22:40.458 -> ===============
19:22:40.468 -> ALL positive score: -7
19:22:40.483 -> ALL negative score: -18
19:22:40.504 -> True Positive
19:22:40.515 ->
19:22:40.521 -> Im020_1.jpg
19:22:43.194 -> ===============
19:22:43.201 -> ALL positive score: -14
19:22:43.223 -> ALL negative score: -6
19:22:43.229 -> False Negative
19:22:43.238 ->
19:22:43.241 -> Im024_1.jpg
19:22:45.916 -> ===============
19:22:45.922 -> ALL positive score: 18
19:22:45.928 -> ALL negative score: 24
19:22:45.938 -> False Negative
19:22:45.946 ->
19:22:45.950 -> Im026_1.jpg
19:22:48.680 -> ===============
19:22:48.685 -> ALL positive score: 27
19:22:48.695 -> ALL negative score: 24
19:22:48.705 -> True Positive
19:22:48.714 ->
19:22:48.719 -> Im028_1.jpg
19:22:51.409 -> ===============
19:22:51.416 -> ALL positive score: 13
19:22:51.427 -> ALL negative score: 18
19:22:51.439 -> False Negative
19:22:51.448 ->
19:22:51.454 -> Im031_1.jpg
19:22:54.138 -> ===============
19:22:54.148 -> ALL positive score: -13
19:22:54.164 -> ALL negative score: -16
19:22:54.179 -> True Positive
19:22:54.183 ->
19:22:54.188 -> Im035_0.jpg
19:22:56.883 -> ===============
19:22:56.890 -> ALL positive score: 12
19:22:56.901 -> ALL negative score: 20
19:22:56.908 -> True Negative
19:22:56.916 ->
19:22:56.921 -> Im041_0.jpg
19:22:59.631 -> ===============
19:22:59.640 -> ALL positive score: 14
19:22:59.653 -> ALL negative score: 6
19:22:59.663 -> False Positive
19:22:59.673 ->
19:22:59.679 -> Im047_0.jpg
19:23:02.365 -> ===============
19:23:02.373 -> ALL positive score: 25
19:23:02.384 -> ALL negative score: 20
19:23:02.393 -> False Positive
19:23:02.399 ->
19:23:02.404 -> Im053_1.jpg
19:23:05.160 -> ===============
19:23:05.174 -> ALL positive score: 39
19:23:05.190 -> ALL negative score: 5
19:23:05.202 -> True Positive
19:23:05.218 ->
19:23:05.223 -> Im057_1.jpg
19:23:07.881 -> ===============
19:23:07.896 -> ALL positive score: 6
19:23:07.912 -> ALL negative score: -1
19:23:07.928 -> True Positive
19:23:07.937 ->
19:23:07.942 -> Im060_1.jpg
19:23:10.618 -> ===============
19:23:10.630 -> ALL positive score: 25
19:23:10.648 -> ALL negative score: 12
19:23:10.661 -> True Positive
19:23:10.667 ->
19:23:10.673 -> Im063_1.jpg
19:23:13.359 -> ===============
19:23:13.368 -> ALL positive score: 23
19:23:13.382 -> ALL negative score: -52
19:23:13.400 -> True Positive
19:23:13.411 ->
19:23:13.417 -> Im069_0.jpg
19:23:16.097 -> ===============
19:23:16.108 -> ALL positive score: -4
19:23:16.129 -> ALL negative score: 34
19:23:16.148 -> True Negative
19:23:16.159 ->
19:23:16.164 -> Im074_0.jpg
19:23:18.812 -> ===============
19:23:18.819 -> ALL positive score: 22
19:23:18.834 -> ALL negative score: 18
19:23:18.850 -> False Positive
19:23:18.861 ->
19:23:18.867 -> Im088_0.jpg
19:23:21.564 -> ===============
19:23:21.575 -> ALL positive score: -21
19:23:21.594 -> ALL negative score: -24
19:23:21.613 -> False Positive
19:23:21.625 ->
19:23:21.630 -> Im095_0.jpg
19:23:24.274 -> ===============
19:23:24.284 -> ALL positive score: -33
19:23:24.302 -> ALL negative score: -38
19:23:24.321 -> False Positive
19:23:24.333 ->
19:23:24.339 -> Im099_0.jpg
19:23:27.014 -> ===============
19:23:27.025 -> ALL positive score: -46
19:23:27.042 -> ALL negative score: -22
19:23:27.062 -> True Negative
19:23:27.074 ->
19:23:27.080 -> Im101_0.jpg
19:23:29.769 -> ===============
19:23:29.779 -> ALL positive score: -17
19:23:29.796 -> ALL negative score: -14
19:23:29.816 -> True Negative
19:23:29.830 ->
19:23:29.837 -> Im106_0.jpg
19:23:32.530 -> ===============
19:23:32.545 -> ALL positive score: -42
19:23:32.562 -> ALL negative score: -45
19:23:32.587 -> False Positive
19:23:32.602 ->
19:23:32.609 -> True Positives: 7
19:23:34.833 -> False Positives: 6
19:23:34.844 -> True Negatives: 4
19:23:34.858 -> False Negatives: 3We see that our model that can correctly classify all twenty images only gets 11/20 when running on Arduino. There are some additional testing steps we can take which will be introduced in V2 that will allow us to test the Arduino model on our development machine to help identify where the bug is coming from. For now this is a good first attempt at building a classifier to detect Acute Lymphoblastic Leukemia detection on Arduino. If you would like to view the ongoing issue in the Tensorflow Micro repository click here, thanks to Advait Jain for the asistance with this issue.
ContributingAsociación de Investigacion en Inteligencia Artificial Para la Leucemia Peter Moss encourages and welcomes code contributions, bug fixes and enhancements from the Github community.
Please read the CONTRIBUTING document for a full guide to forking our repositories and submitting your pull requests. You will also find our code of conduct in the Code of Conduct document.
Contributors- Adam Milton-Barker - Asociación de Investigacion en Inteligencia Artificial Para la Leucemia Peter Moss President/Founder & Lead Developer, Sabadell, Spain
We use SemVer for versioning.
LicenseThis project is licensed under the MIT License - see the LICENSE file for details.
Bugs/IssuesWe use the repo issues to track bugs and general requests related to using this project. See CONTRIBUTING for more info on how to submit bugs, feature requests and proposals.















Comments
Please log in or sign up to comment.