According to the Skin Cancer Foundation, half of the population in United States are diagnosed with some form of skin cancer by age 65. The survival rate for early detection is almost 98%, but it falls to 62% when the cancer reaches the lymph node and 18% when it metastasizes to distance organs. With Skin Cancer AI, we want to use power of artificial intelligence to provide early detection as widely as available.
Deep learning has been a pretty big trend for machine learning lately, and the recent success has paved the way to build project like this. We are going to focus specifically on computer vision and image classification in this sample. To do this, we will be building nevus, melanoma, and seborrheic keratosis image classifier using deep learning algorithm, the Convolution Neural Network (CNN) through Caffe Framework.
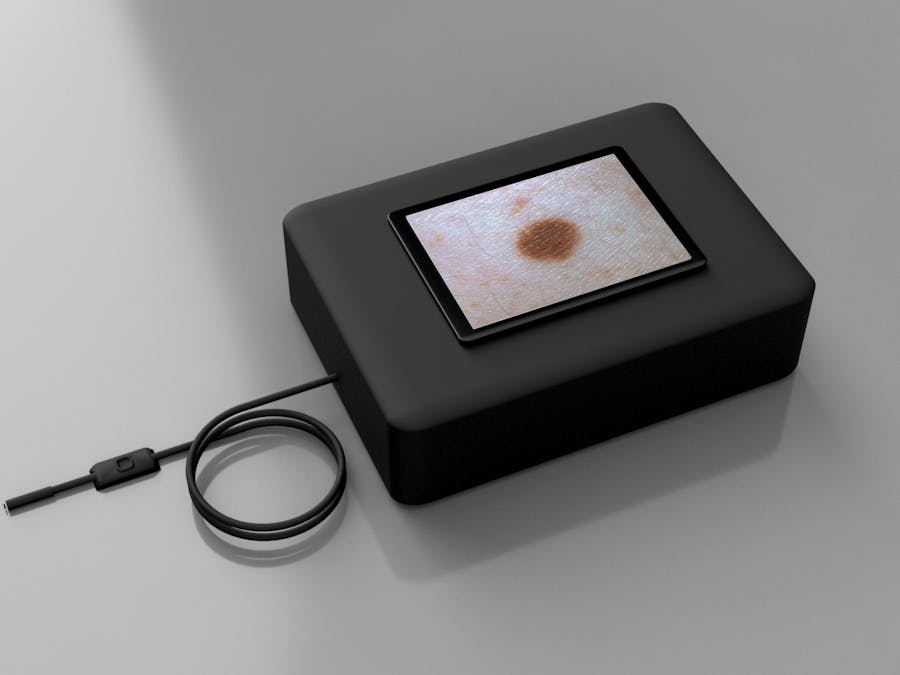
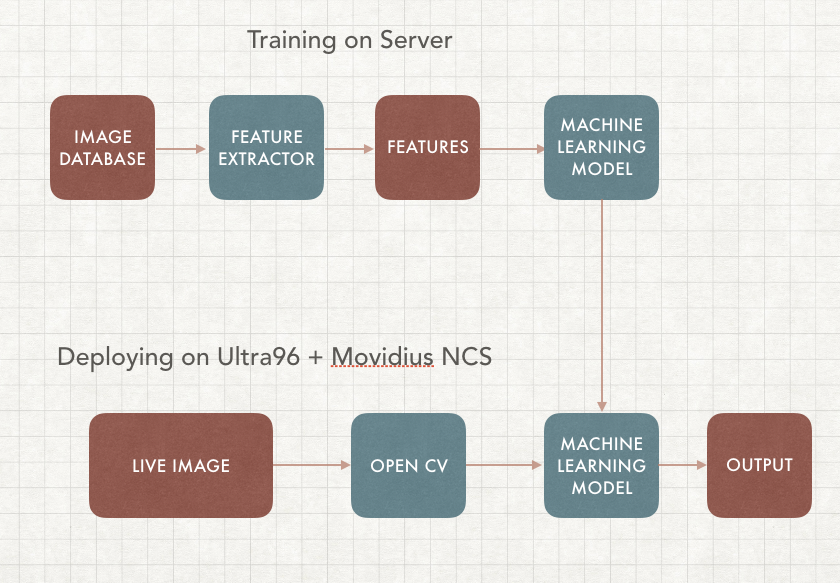
In this article we will focus on Supervised learning, it requires training on the server as well as deploying on the edge. Our goal is to build a machine learning algorithm that can detect cancer images in real time, this way you can build your own AI based skin cancer classification device.
Our application will include 2 parts, the first part is training, which we will be using different sets of cancer image database to train a machine learning algorithm (model) with their corresponding labels. The second part is deploying on the edge, which uses the same model we've trained and running it on an Edge device, in this case Movidius Neural Computing Stick through Ultra96 FPGA. This way VPU can run inference while FPGA can do the OpenCV
Traditional Machine Learning (ML) vs. Deep LearningThis is probably the most asked question in AI, and it's fairly simple once you learn how to do it. In order to understand this we first have to learn how machine learning image classification works.
Machine learning requires feature extraction and model training. We first have to use domain knowledge to extract features that can be used for our ML algorithm model, some examples includes SIFT and HoG. After that we can use a dataset that has all the image features and labels to train our machine learning model.
The major difference between traditional ML and Deep Learning is in the feature engineering. The traditional ML uses manually programmed features where Deep Learning does it automatically. Feature engineering is relatively difficult since it requires domain expertise and is very time consuming. Deep learning requires no feature engineering and can be more accurate
Artificial Neural Networks (ANNs)
According to techopedia, "An artificial neuron network (ANN) is a computational model based on the structure and functions of biological neural networks." The artificial neuron network is technically emulating how human biological neuron works, it has a finite number of inputs, weights associate with them, and an activate function. The activation function of a node defines the output of that node given an input or set of inputs, It is non-linear to encode complex. patterns of the data. When input comes in, the activation function applies to the weight sum of the inputs to generate the output. The artificial neurons are connected to one another to form a network, hence it's called artificial neuron network (ANN).
A feedforward neural network is an artificial neural network wherein connections between the nodes do not form a cycle, this is the simplest form of ANN. It has 3 layers, Input, Hidden and Output layer, where the data comes in through Input Layer, through the hidden layer onto the output nodes like the figure below. We can have multiple hidden layers, the complexity of the model is correlated to the size of hidden layers.
Training data an Loss function are the 2 elements used in training a neural network. Training data is composed of images and the corresponding labels; and Loss function is a function that measures the inaccuracies during classification. Once obtained those 2 elements, we then use backpropagation algorithm and gradient descent to train an ANN.
Convolutional Neural Networks (CNNs)
Convolutional neural network is a class of deep, feed-forward artificial neural networks, most commonly applied to analyzing visual imagery because it is designed to emulate behavior of biological behaviors on animal visual cortex. It consist of convolutional layers and pooling layers so that the network can encode image properties.
The convolutional layer's parameters consists of a set of learnable filters (or kernels) that have a small receptive field. This way the image can convolve across spatially, and computing dot products between the entries of the filter and the input and producing a 2-dimensional activation map of that filter. This way the network learns filters that can activate when it detects special features on the input image's spatial feature.
The pooling layer is a form of non-linear down-sampling. It partitions the input image into a set of non-overlapping rectangles and, for each such sub-region, outputs the maximum. The idea is to continuously reduce the spatial size of the input representation to reduce the amount of parameters and computation in the network, so it can also control overfitting. Max pooling is the most common type non-linear pooling. According to wikipedia, "Pooling is often applied with filters of size 2x2 applied with a stride of 2 at every depth slice. A pooling layer of size 2x2 with stride of 2 shrinks the input image to a 1/4 of its original size."
Equipments needed is very simple for this project, you can either do it with your computer and a USB Movidius Neural Computing Stick and Ultra96 board.
- Ultra96 Board
- Endoscope Camera
- Movidius Neural Computing Stick
- A screen or monitor
Ultra96 is pretty new, but the support group was kind enough to get the base Ubuntu running, which is a great deal since this allows me to build different platforms off ultra96. The compiled version of debian can be downloaded fromhttps://fileserver.linaro.org/owncloud/index.php/s/jTt3MYSuwtLuf9d After that we can use tools like ether to load it onto the mini-sd card.
Upon booting, we first have to fix some errors by removing corrupted repos.
sudo rm -r /var/lib/apt/lists/*
This allows us to install all the packages as needed to use the platform.
So to get AI and Computer Vision to work, we can utilize the Movidius NCS which have set of tools to get our project running. There isn't a walkthrough for Ultra96 so we've basically referenced this through https://movidius.github.io/blog/ncs-apps-on-rpi/
First we have to install dependencies, this part does not come with PYNQ, so we will install them to make sure everything works.
apt-get install libgstreamer1.0-0 gstreamer1.0-plugins-base gstreamer1.0-plugins-good gstreamer1.0-plugins-bad gstreamer1.0-plugins-ugly gstreamer1.0-libav gstreamer1.0-doc gstreamer1.0-tools libgstreamer-plugins-base1.0-dev
apt-get install libgtk-3-dev
apt-get install -y libprotobuf-dev libleveldb-dev libsnappy-dev
apt-get install -y libopencv-dev libhdf5-serial-dev
apt-get install -y protobuf-compiler byacc libgflags-dev
apt-get install -y libgoogle-glog-dev liblmdb-dev libxslt-dev
Next we can install NCSDK SDK, which contains API that can connect applications to the NCS. Since NCSDK was not built for Ultra96, we can make following modifications for the workaround with markjay4k's version
cd /home/xilinx/
mkdir -p workspace
cd workspace
git clone https://github.com/markjay4k/ncsdk-aarch64.git
cd ncsdk/api/src
make
make install
This work around should get us the NCSDK API working with Ultra96 and PYNQ
Next we will try to get NcAppZoo and Hello World running for Neural Computing Stick, we need to repeat the old python 3.6 on pynq
cd /home/xilinx/workspace
git clone https://github.com/movidius/ncappzoo
cd ncappzoo/apps/hello_ncs_py
make run
And you just got NCS to run on aarch64 :)
There are a lot of ways we can make the trigger, since this is an AI based application I will make an AI based trigger. In this guide we will be using a SSD neural net that is pre-trained and working with Caffe, and the detection will be garbage. So we'd also learn how to utilize other neural network with little bit of work.
In this step we, previously we've trained the through caffe model, we have to compile the graph on another machine as we've only installed API and not toolkit as they are not being built for aarch64. However, since the API works, we can simply build it on another machine and transfer the graph file over to the Ultra96. The FPGA can handle all the CV2, and the Movidius NCS here will be running inferencing for object detection and image classification as image below, we are going determine the basics by doing live image classification here.
We first need skin cancer data sets, although there are many places to get it, isic-archive becomes the easiest one. For this part we just need about 500 images of each between nevus, melanoma and seborrheic keratosis as well as 500 of random images of anything else. To get higher accuracy we'd have to use more data, this is just to get training started. Easiest way to get the data is via image below using https://isic-archive.com/#images
After wards we can tag the images into one folder, and naming them nevus-00.jpg, melanoma-00.jpg so the we can form our lmdb easily.
Machine learning training uses a lot of processing power, and hence they generally cost a lot. In this article we are going to focus on AI DevCloud which is free for Intel AI Academy members.
To build Skin Cancer AI, we are using Caffe Framework, mainly because of the Intel Movidius NCS's AI on the Edge support. Caffe is a deep learning framework developed by the Berkeley Vision and Learning Center (BVLC), and there are 4 steps of using caffe framework to train our algorithm model. This way Xilinx' FPGA can focus on openCV and NCS can focus on inferencing.
- Data Prep, we clean the images and store them into LMDB.
- Model Definition prototxt file, define the parameters and choose CNN architecture.
- Solver Definition prototxt file, define the solver parameters for model optimization
- Model training, we execute caffe command to get our .caffemodel algorithm file
On Devcloud, we can check whether these are available simply by going to
cd /glob/deep-learning/py-faster-rcnn/caffe-fast-rcnn/build/tools
Once we have DevCloud setup, we can build it up
mkdir skincancerai
cd skincancerai
mkdir input
cd input
mkdir train
And from there we can get all the data we've previously setup into the folder
scp ./* colfax:/home/[youruser_name]/skincancerai/input/train/
After that we can build our lmdb this way. And we do it through following sets
- 5/6 of the data set will be used for training, 1/6 is being used for validation so we can calculate the accuracy of the model
- We are resizing all images to 227x227 to follow the same standard as BVLC
- Histogram equalization is being applied to all the training images to adjust the contrast.
- And store them among train_lmdb and validation_lmdb
- Use make_datum to label all the image dataset inside lmdb
import os
import glob
import random
import numpy as np
import cv2
import caffe
from caffe.proto import caffe_pb2
import lmdb
#We use 227x227 from BVLC
IMAGE_WIDTH = 227
IMAGE_HEIGHT = 227
def transform_img(img, img_width=IMAGE_WIDTH, img_height=IMAGE_HEIGHT):
img[:, :, 0] = cv2.equalizeHist(img[:, :, 0])
img[:, :, 1] = cv2.equalizeHist(img[:, :, 1])
img[:, :, 2] = cv2.equalizeHist(img[:, :, 2])
img = cv2.resize(img, (img_width, img_height), interpolation = cv2.INTER_CUBIC)
return img
def make_datum(img, label):
return caffe_pb2.Datum(
channels=3,
width=IMAGE_WIDTH,
height=IMAGE_HEIGHT,
label=label,
data=np.rollaxis(img, 2).tostring())
train_lmdb = '/home/[your_username]/skincancerai/input/train_lmdb'
validation_lmdb = '/home/[youser_username]/skincancerai/input/validation_lmdb'
os.system('rm -rf ' + train_lmdb)
os.system('rm -rf ' + validation_lmdb)
train_data = [img for img in glob.glob("./input/train/*jpg")]
random.shuffle(train_data)
print 'Creating train_lmdb'
in_db = lmdb.open(train_lmdb, map_size=int(1e12))
with in_db.begin(write=True) as in_txn:
for in_idx, img_path in enumerate(train_data):
if in_idx % 6 == 0:
continue
img = cv2.imread(img_path, cv2.IMREAD_COLOR)
img = transform_img(img, img_width=IMAGE_WIDTH, img_height=IMAGE_HEIGHT)
if 'none' in img_path:
label = 0
elif 'nevus' in img_path:
label = 1
elif 'melanoma' in img_path:
label = 2
else:
label = 3
datum = make_datum(img, label)
in_txn.put('{:0>5d}'.format(in_idx), datum.SerializeToString())
print '{:0>5d}'.format(in_idx) + ':' + img_path
in_db.close()
print '\nCreating validation_lmdb'
in_db = lmdb.open(validation_lmdb, map_size=int(1e12))
with in_db.begin(write=True) as in_txn:
for in_idx, img_path in enumerate(train_data):
if in_idx % 6 != 0:
continue
img = cv2.imread(img_path, cv2.IMREAD_COLOR)
img = transform_img(img, img_width=IMAGE_WIDTH, img_height=IMAGE_HEIGHT)
if 'none' in img_path:
label = 0
elif 'nevus' in img_path:
label = 1
elif 'melanoma' in img_path:
label = 2
else:
label = 3
datum = make_datum(img, label)
in_txn.put('{:0>5d}'.format(in_idx), datum.SerializeToString())
print '{:0>5d}'.format(in_idx) + ':' + img_path
in_db.close()
print '\nFinished processing all images'
After wards we will run the script
python2 create_lmdb.py
to get all the LMDB. After that is done, we will need to get the mean image of the training data. As part of the caffe, we can do it through
cd /glob/deep-learning/py-faster-rcnn/caffe-fast-rcnn/build/toolscompute_image_mean -backend=lmdb /home/[your_user]/skincancerai/input/train_lmdb /home/[your_user]/skincancerai/input/mean.binaryproto
The command above will generate the mean image of training data. Each input image will subtract the mean image so that every feature pixel has zero mean. This is a commonly used in preprocessing for supervised machine learning.
Step 7: Set Up Model Definition and Solver DefinitionWe now need to setup model definition and solver defination, in this article we will be using bvlc_reference_net, which can be seen at https://github.com/BVLC/caffe/tree/master/models/bvlc_reference_caffenet
Below is modified version of train.prototxt
name: "CaffeNet"
layer {
name: "data"
type: "Data"
top: "data"
top: "label"
include {
phase: TRAIN
}
transform_param {
mirror: true
crop_size: 227
mean_file: "/home/[your_username]/skincancerai/input/mean.binaryproto"
}
data_param {
source: "/home/[your_username]/skincancerai/input/train_lmdb"
batch_size: 128
backend: LMDB
}
}
layer {
name: "data"
type: "Data"
top: "data"
top: "label"
include {
phase: TEST
}
transform_param {
mirror: false
crop_size: 227
mean_file: "/home/[your_username]/skincancerai/input/mean.binaryproto"
}
# mean pixel / channel-wise mean instead of mean image
# transform_param {
# crop_size: 227
# mean_value: 104
# mean_value: 117
# mean_value: 123
# mirror: true
# }
data_param {
source: "/home/[your_username]/skincancerai/input/validation_lmdb"
batch_size: 36
backend: LMDB
}
}
layer {
name: "conv1"
type: "Convolution"
bottom: "data"
top: "conv1"
param {
lr_mult: 1
decay_mult: 1
}
param {
lr_mult: 2
decay_mult: 0
}
convolution_param {
num_output: 96
kernel_size: 11
stride: 4
weight_filler {
type: "gaussian"
std: 0.01
}
bias_filler {
type: "constant"
value: 0
}
}
}
layer {
name: "relu1"
type: "ReLU"
bottom: "conv1"
top: "conv1"
}
layer {
name: "pool1"
type: "Pooling"
bottom: "conv1"
top: "pool1"
pooling_param {
pool: MAX
kernel_size: 3
stride: 2
}
}
layer {
name: "norm1"
type: "LRN"
bottom: "pool1"
top: "norm1"
lrn_param {
local_size: 5
alpha: 0.0001
beta: 0.75
}
}
layer {
name: "conv2"
type: "Convolution"
bottom: "norm1"
top: "conv2"
param {
lr_mult: 1
decay_mult: 1
}
param {
lr_mult: 2
decay_mult: 0
}
convolution_param {
num_output: 256
pad: 2
kernel_size: 5
group: 2
weight_filler {
type: "gaussian"
std: 0.01
}
bias_filler {
type: "constant"
value: 1
}
}
}
layer {
name: "relu2"
type: "ReLU"
bottom: "conv2"
top: "conv2"
}
layer {
name: "pool2"
type: "Pooling"
bottom: "conv2"
top: "pool2"
pooling_param {
pool: MAX
kernel_size: 3
stride: 2
}
}
layer {
name: "norm2"
type: "LRN"
bottom: "pool2"
top: "norm2"
lrn_param {
local_size: 5
alpha: 0.0001
beta: 0.75
}
}
layer {
name: "conv3"
type: "Convolution"
bottom: "norm2"
top: "conv3"
param {
lr_mult: 1
decay_mult: 1
}
param {
lr_mult: 2
decay_mult: 0
}
convolution_param {
num_output: 384
pad: 1
kernel_size: 3
weight_filler {
type: "gaussian"
std: 0.01
}
bias_filler {
type: "constant"
value: 0
}
}
}
layer {
name: "relu3"
type: "ReLU"
bottom: "conv3"
top: "conv3"
}
layer {
name: "conv4"
type: "Convolution"
bottom: "conv3"
top: "conv4"
param {
lr_mult: 1
decay_mult: 1
}
param {
lr_mult: 2
decay_mult: 0
}
convolution_param {
num_output: 384
pad: 1
kernel_size: 3
group: 2
weight_filler {
type: "gaussian"
std: 0.01
}
bias_filler {
type: "constant"
value: 1
}
}
}
layer {
name: "relu4"
type: "ReLU"
bottom: "conv4"
top: "conv4"
}
layer {
name: "conv5"
type: "Convolution"
bottom: "conv4"
top: "conv5"
param {
lr_mult: 1
decay_mult: 1
}
param {
lr_mult: 2
decay_mult: 0
}
convolution_param {
num_output: 256
pad: 1
kernel_size: 3
group: 2
weight_filler {
type: "gaussian"
std: 0.01
}
bias_filler {
type: "constant"
value: 1
}
}
}
layer {
name: "relu5"
type: "ReLU"
bottom: "conv5"
top: "conv5"
}
layer {
name: "pool5"
type: "Pooling"
bottom: "conv5"
top: "pool5"
pooling_param {
pool: MAX
kernel_size: 3
stride: 2
}
}
layer {
name: "fc6"
type: "InnerProduct"
bottom: "pool5"
top: "fc6"
param {
lr_mult: 1
decay_mult: 1
}
param {
lr_mult: 2
decay_mult: 0
}
inner_product_param {
num_output: 4096
weight_filler {
type: "gaussian"
std: 0.005
}
bias_filler {
type: "constant"
value: 1
}
}
}
layer {
name: "relu6"
type: "ReLU"
bottom: "fc6"
top: "fc6"
}
layer {
name: "drop6"
type: "Dropout"
bottom: "fc6"
top: "fc6"
dropout_param {
dropout_ratio: 0.5
}
}
layer {
name: "fc7"
type: "InnerProduct"
bottom: "fc6"
top: "fc7"
param {
lr_mult: 1
decay_mult: 1
}
param {
lr_mult: 2
decay_mult: 0
}
inner_product_param {
num_output: 4096
weight_filler {
type: "gaussian"
std: 0.005
}
bias_filler {
type: "constant"
value: 1
}
}
}
layer {
name: "relu7"
type: "ReLU"
bottom: "fc7"
top: "fc7"
}
layer {
name: "drop7"
type: "Dropout"
bottom: "fc7"
top: "fc7"
dropout_param {
dropout_ratio: 0.5
}
}
layer {
name: "fc8"
type: "InnerProduct"
bottom: "fc7"
top: "fc8"
param {
lr_mult: 1
decay_mult: 1
}
param {
lr_mult: 2
decay_mult: 0
}
inner_product_param {
num_output: 4
weight_filler {
type: "gaussian"
std: 0.01
}
bias_filler {
type: "constant"
value: 0
}
}
}
layer {
name: "accuracy"
type: "Accuracy"
bottom: "fc8"
bottom: "label"
top: "accuracy"
include {
phase: TEST
}
}
layer {
name: "loss"
type: "SoftmaxWithLoss"
bottom: "fc8"
bottom: "label"
top: "loss"
}
At the same time we can create deploy.prototxt which is built off train.prototxt. This can be seen from the github repo. We will also create the label.txt file the same way we created the lmdb file
classes
None
Nevus
Melanoma
Seborrheic Keratosis
After that we need Solver Definition in solver.prototxt, it is used to optimize the training models. Because we are relying on CPU, we need to make some modifications on the solver definition below.
net: "/home/[your_username]/skincancerai/model/train.prototxt"
test_iter: 50
test_interval: 50
base_lr: 0.001
lr_policy: "step"
gamma: 0.1
stepsize: 50
display: 50
max_iter: 5000
momentum: 0.9
weight_decay: 0.0005
snapshot: 1000
snapshot_prefix: "/home/[your_username]/skincancerai/model"
solver_mode: CPU
Because we are dealing with small amount of data here, we can shorten the test iterations and get our model as quick as possible. To make this short, the solver will compute the accuracy of the model every 50 iterations using the validation set. Since we don't have a lot of data, the solver optimization process will take a snapshot every 1000 iteration and run for a maximum of 5000 iterations. Current configuration of lr_policy: "step", stepsize: 2500, base_lr: 0.001 and gamma: 0.1 is pretty standard as we can try to use others as well through bvlc solver documentation.
Step 8: Training the ModelSince we are using the free AI DevCloud and have everything all set, we can use the Intel Caffe which is optimized using Intel CPU that's installed on the cluster. Since this is a cluster, we can simply start training by using the command below.
cd /glob/deep-learning/py-faster-rcnn/caffe-fast-rcnn/build/toolsecho caffe train --solver ~/skincancerai/model/solver.prototxt | qsub -o ~/skincancerai/model/output.txt -e ~/skincancerai/model/train.log
The trained model will be model_iter_1000.caffemodel,model_iter_2000.caffemodel and so on. With the data given from ISIC you should obtain somewhere around 70 to 80% accuracy. You can plot your own curve by the command below,
cd ~/skincancerai python2 plot_learning_curve.py ./model/train.log ./model/train.png
For this article we are using Ultra96 so that we can have an offline device which we can carry around everywhere. We've pretty much got the sample running on Step 3. Since we do not have toolkit active on aarch64, we can run the compilation on another machine and then copy it into ultra96
mvNCCompile deploy.prototxt -w
model_iter_1000.caffemodel,model_iter_2000.caffemodel
This would give you the graph file, copy the graph file and the label file and put it under CancerNet along with following code that we modified, copy it onto Ultra96. We now have a skin cancer detection device that can be used at home. We can deploy following code with graph file and categories.txt file under CancerNet
#!/usr/bin/python3
# ****************************************************************************
# Copyright(c) 2017 Intel Corporation.
# License: MIT See LICENSE file in root directory.
# ****************************************************************************
# Perform inference on a LIVE camera feed using DNNs on
# Intel® Movidius™ Neural Compute Stick (NCS)
import os
import cv2
import sys
import numpy
import ntpath
import argparse
import mvnc.mvncapi as mvnc
# Variable to store commandline arguments
ARGS = None
# OpenCV object for video capture
camera = None
# ---- Step 1: Open the enumerated device and get a handle to it -------------
def open_ncs_device():
# Look for enumerated NCS device(s); quit program if none found.
devices = mvnc.EnumerateDevices()
if len( devices ) == 0:
print( "No devices found" )
quit()
# Get a handle to the first enumerated device and open it
device = mvnc.Device( devices[0] )
device.OpenDevice()
return device
# ---- Step 2: Load a graph file onto the NCS device -------------------------
def load_graph( device ):
# Read the graph file into a buffer
with open( ARGS.graph, mode='rb' ) as f:
blob = f.read()
# Load the graph buffer into the NCS
graph = device.AllocateGraph( blob )
return graph
# ---- Step 3: Pre-process the images ----------------------------------------
def pre_process_image( frame ):
# Resize image [Image size is defined by choosen network, during training]
img = cv2.resize( frame, tuple( ARGS.dim ) )
# Extract/crop a section of the frame and resize it
height, width, channels = frame.shape
x1 = int( width / 3 )
y1 = int( height / 4 )
x2 = int( width * 2 / 3 )
y2 = int( height * 3 / 4 )
cv2.rectangle( frame, ( x1, y1 ) , ( x2, y2 ), ( 0, 255, 0 ), 2 )
img = frame[ y1 : y2, x1 : x2 ]
# Resize image [Image size if defined by choosen network, during training]
img = cv2.resize( img, tuple( ARGS.dim ) )
# Convert BGR to RGB [OpenCV reads image in BGR, some networks may need RGB]
if( ARGS.colormode == "rgb" ):
img = img[:, :, ::-1]
# Mean subtraction & scaling [A common technique used to center the data]
img = img.astype( numpy.float16 )
img = ( img - numpy.float16( ARGS.mean ) ) * ARGS.scale
return img
# ---- Step 4: Read & print inference results from the NCS -------------------
def infer_image( graph, img, frame ):
# Load the image as a half-precision floating point array
graph.LoadTensor( img, 'user object' )
# Get the results from NCS
output, userobj = graph.GetResult()
# Find the index of highest confidence
top_prediction = output.argmax()
# Get execution time
inference_time = graph.GetGraphOption( mvnc.GraphOption.TIME_TAKEN )
print( "I am %3.1f%%" % (100.0 * output[top_prediction] ) + " confidant"
+ " it is " + labels[top_prediction]
+ " ( %.2f ms )" % ( numpy.sum( inference_time ) ) )
displaystring = str(100.0 * output[top_prediction]) + " " + labels[top_prediction]
# If a display is available, show the image on which inference was performed
if 'DISPLAY' in os.environ:
textsize = cv2.getTextSize(displaystring, cv2.FONT_HERSHEY_SIMPLEX, 1, 2)[0]
textX = (frame.shape[1] - textsize[0])/2
cv2.putText(frame, displaystring, (int(textX),450), cv2.FONT_HERSHEY_SIMPLEX, 1, (255,0,0),2)
cv2.imshow( 'Skin Cancer AI', frame )
# ---- Step 5: Unload the graph and close the device -------------------------
def close_ncs_device( device, graph ):
graph.DeallocateGraph()
device.CloseDevice()
camera.release()
cv2.destroyAllWindows()
# ---- Main function (entry point for this script ) --------------------------
def main():
device = open_ncs_device()
graph = load_graph( device )
# Main loop: Capture live stream & send frames to NCS
while( True ):
ret, frame = camera.read()
img = pre_process_image( frame )
infer_image( graph, img, frame )
# Display the frame for 5ms, and close the window so that the next
# frame can be displayed. Close the window if 'q' or 'Q' is pressed.
if( cv2.waitKey( 5 ) & 0xFF == ord( 'q' ) ):
break
close_ncs_device( device, graph )
# ---- Define 'main' function as the entry point for this script -------------
if __name__ == '__main__':
parser = argparse.ArgumentParser(
description="Image classifier using \
Intel® Movidius™ Neural Compute Stick." )
parser.add_argument( '-g', '--graph', type=str,
default='CancerNet/graph',
help="Absolute path to the neural network graph file." )
parser.add_argument( '-v', '--video', type=int,
default=0,
help="Index of your computer's V4L2 video device. \
ex. 0 for /dev/video0" )
parser.add_argument( '-l', '--labels', type=str,
default='./CancerNet/categories.txt',
help="Absolute path to labels file." )
parser.add_argument( '-M', '--mean', type=float,
nargs='+',
default=[78.42633776, 87.76891437, 114.89584775],
help="',' delimited floating point values for image mean." )
parser.add_argument( '-S', '--scale', type=float,
default=1,
help="Absolute path to labels file." )
parser.add_argument( '-D', '--dim', type=int,
nargs='+',
default=[227, 227],
help="Image dimensions. ex. -D 224 224" )
parser.add_argument( '-c', '--colormode', type=str,
default="rgb",
help="RGB vs BGR color sequence. This is network dependent." )
ARGS = parser.parse_args()
# Create a VideoCapture object
camera = cv2.VideoCapture( ARGS.video )
# Set camera resolution
camera.set( cv2.CAP_PROP_FRAME_WIDTH, 620 )
camera.set( cv2.CAP_PROP_FRAME_HEIGHT, 480 )
# Load the labels file
labels =[ line.rstrip('\n') for line in
open( ARGS.labels ) if line != 'classes\n']
main()
# ==== End of file ===========================================================
And from here, we have our Skin Cancer AI classification, simply run the following command in the folder.
python3 live-image-classifier.py
And we can detect our own moles
And when there is melanoma
And we can see the full demo

















Comments
Please log in or sign up to comment.