The plant diseases are a major thread to losses of modern agricultural production. Plant disease severity is an important parameter to measure disease level and thus can be used to predict yield and recommend treatment. The rapid, accurate diagnosis of disease severity will help to reduce yield losses. Traditionally, plant disease severity is scored with visual inspection of plant tissue by trained experts. The expensive cost and low efficiency of human disease assessment hinder the rapid development of modern agriculture. With the population of digital cameras and the advances in computer vision, the automated disease diagnosis models are highly demanded by precision agriculture, high-throughput plant phenotype, smart green house, and so forth.
Apples may be grown in many parts of the country and lend themselves well to part-time farming operations. The initial investment for apples can be high depending on the production method chosen, land preparation, and initial investment in the trees. A commercial orchard is expected to be productive for at least 20 years, so this investment will be spread over a longer period of time than many crops. Depending on the amount of land devoted to the orchard, production method, and tree size, equipment costs may be held to a minimum. If the orchard is a part of an existing agricultural operation, you may already have much of the needed equipment.
Apple production will require many hours of labor, depending on the size of the orchard. Land preparation and planting will require at least two people. During the summer months, the orchard will require mowing, multiple pesticide applications, and fruit thinning. Depending on the mix of varieties and orchard size, additional labor may be required at harvest time.
According to the United States Department of Agriculture's (USDA) National Agricultural Statistics Service, more than 5, 000 farms with almost 85, 000 acres of apples are located in the northeastern United States. Pennsylvania produces 400 to 500 million pounds of apples per year and ranks fourth in the nation for apple production.
Agriculture is the primary occupation of villagers and a large sustaining population is dependent on agriculture.From the advent of agriculture, there has been much mechanical and chemical advancement that has occurred to improve the yield and help farmers tackle issues like agriculture and crop diseases. But there has been little to less digitization done in this field. With the boom of IOT, there is a hope for creating a digital system for agriculture which will help the farmer make informed decisions about his farm and help him tackle some undesired situations in advance. So, it will help to improve the quality of crops and also it will be beneficial for farmers. Early Detection of Disease which is a great challenge in agriculture field. An earlier large team of experts are called by the farmers to chalk out the diseases or any harm which occurred to plants, even this practice is not known to every farmer and therefore the experts cost much and also it is time consuming. Whereas Automatic detection is more beneficial than this long process of observations by the experts, Automation technique of the disease detection where the result comes out to just monitoring the change in plant.
Conclusion🍎:Creating Hydra: OpenVino based Plant Disease and climatic factors monitoring and autonomous watering system.
Taking into consideration these drawbacks faced during manual crop monitoring, I decided to create an autonomous monitoring system. In manual methods of agriculture, due to insufficient crop data, there is inaccurate crop monitoring, This leads to low crop yield production. In this project, I've taken into consideration the low yielding of the apple production across the world and based on this crop factorial conditions, I have designed the logic. In Apple plant production, stability of constant soil PH levels are required along with constant temperature and humidity for accurate high yield productions. Depending on the Diseases faced by the Apple plant, the plant array has to be provided with the suitable conditions. For example: If an Apple plant disease is detected where in the calcium levels of the plant is low, the plant has to be provided with the similar nutrients for which it is facing deficiency.
Based on this conclusion, I decided to create a Vision Learning model which accuratly detects the diseases faced in Apple plantations and provide geo spatial analysis of this data across the farm along with Timely Data Trends.
The below Pie Chart potrays the most observed diseases in Apple Plantations🍏:
This Pie chart displays the percentage-wise distribution of commonly faced diseases in Apple Agriculture.
Taking into consideration these commonly faced diseases, I decided to create a Computer Vision Model based on OpenVino deployed on the Raspberry Pi for detection and classification of these diseases.
For this purpose, I have taken into consideration the following most commonly faced Diseases:
1)Alternaria Leaf Blotch:
Lesions first appear on leaves in late spring or early summer as small, round, purplish or blackish spots, gradually enlarging to 1/8 to ¼ inch in diameter and have a purple border. Some spots turn grayish brown, but most lesions may coalesce or undergo secondary enlargement and become irregular and much darker, acquiring a "frog-eye" appearance. When lesions occur on petioles, the leaves turn yellow and 50 percent or more defoliation may occur. Severe defoliation leads to premature fruit drop. Fruit infections result in small, dark, raised lesions associated with the lenticel. Alternaria leaf blotch is most likely to occur on 'Delicious' strains and should not be confused with frogeye leaf spot, captan spot, or with 'Golden Delicious' necrotic leaf blotch. Frogeye leaf spot usually appears earlier in the season and is associated with nearby dead wood or fruit mummies. Captan spot spray injury occurs when captan fungicide is applied under wet conditions and associated with 2 to 4 leaves on terminals, representing a spray event. 'Golden Delicious' necrotic leaf blotch commonly occurs in July and August as a result of physiological stress caused by fluctuating soil moisture. Alternaria leaf blotch tends to be uniformly distributed throughout the tree.
2) Cedar Apple Rust:
Cedar-apple rust is the most common of the three fungal rust diseases and attacks susceptible cultivars of apples and crabapples. It infects the leaves, fruit, and, occasionally, young twigs. The alternate host plant, Eastern red cedar (Juniperus virginiana), is necessary for the survival of the fungus.
3) Fire Blight:
Fire blight is a common and very destructive bacterial disease of apples and pears (Figure 1). The disease is caused by the bacterium Erwinia amylovora, which can infect and cause severe damage to many plants in the rose (Rosaceae) family (Table 1). On apples and pears, the disease can kill blossoms, fruit, shoots, twigs, branches and entire trees. While young trees can be killed in a single season, older trees can survive several years, even with continuous dieback.
4) Powdery Mildew of apples:
Powdery mildew of apples, caused by the fungus Podosphaera leucotricha, forms a dense white fungal growth (mycelium) on the host tissue and affects:1)leaves2)buds3)shoots4)fruits.The disease stunts the growth of trees and is found wherever apples are grown. It's also been reported on pear and quince, though damage on these hosts is rarely seen in Australia.
5) Apple Leaf Rollers:
Pest description and crop damage Several species of leafrollers [family: Tortricidae] are pests of tree fruits. These species use native host plants as well as fruit trees. The different species of leafroller cause similar damage to apple trees but differ in appearance and life cycle. The principal leafroller pests of fruit trees can be divided into single-generation moths, such as the fruittree leafroller and the European leafroller, and two-generation moths, such as the obliquebanded leafroller and pandemis leafroller.
Using the above set of most commonly faced diseases in Apple plants, I have trained a computer vision Tensorflow Model that helps in pre prediction of diseases in apple plants to prevent loss of Crop yield.
The Disease prediction and detection computer vision model was meant to detect diseases in Apple Plants. Besides this, plant and soil health monitoring along with Climate Tracking Models are essential for analysis for Apple plants.
The Hydra Tool consists of a framework which analyses the following models to perform inference and provide visual Analysis of the Data:
- Computer Vision OpenVino optimized Model for Plant disease Detection and Classification
- Soil Moisture Model for tracking water level in soils and maintaining water level threshold by autonomous Watering using peristaltic pump.
- Temperature and Humidity Model to keep in track Climatic and Environmental factors suitable for a high yield since Apple Plants need precise and stable climatic condition for constant growth.
- Front-end Kepler.gl tool for geospatial analysis of the farm using Satellite Imaging and Apple Plant location wise Individual sensor data plotting.
- Front-end Streamlit tool for Mean Average Apple crop disease Analysis and manual Soil moisture level maintaining through commands sent to peristaltic pump for watering.
1) Raspberry Pi 4 model B (2Gb/4Gb)
The quad-core Raspberry Pi 4 Model B is both faster and more capable than its predecessor, the Raspberry Pi 3 Model B+. For those interested in benchmarks, the Pi 4's CPU -- the board's main processor -- is offering two to three times the performance of the Pi 3's processor in some benchmarks. The quad-core Raspberry Pi 4 Model B is both faster and more capable than its predecessor, the Raspberry Pi 3 Model B+. For those interested in benchmarks, the Pi 4's CPU -- the board's main processor -- is offering two to three times the performance of the Pi 3's processor in some benchmarks. Unlike its predecessor, the new board is capable of playing 4K video at 60 frames per second, boosting the Pi's media center credentials.
Connecting the Raspberry Pi Camera:
2)Intel Neural Compute Stick 2
Intel Neural Compute Stick 2 is powered by the Intel Movidius X VPU to deliver industry-leading performance, wattage, and power. The NEURAL COMPUTE supports OpenVINO, a toolkit that accelerates solution development and streamlines deployment. The Neural Compute Stick 2 offers plug-and-play simplicity, support for common frameworks and out-of-the-box sample applications. Use any platform with a USB port to prototype and operate without cloud compute dependence. The Intel NCS 2 delivers 4 trillion operations per second with an 8X performance boost compared to previous generations.
3) Raspberry Pi Camera Module:
It's capable of 3280 x 2464 pixel static images, and also supports 1080p30, 720p60 and 640x480p90 video.It attaches to the Raspberry Pi via one of the small sockets on the board's upper surface and uses the dedicated CSi interface, designed especially for interfacing to cameras.Features:
- Supports 1080p30, 720p60 and 640x480p90 videoSize 25mm x 23mm x 9mmWeight just over 3g
- Supports 1080p30, 720p60 and 640x480p90 videoSize 25mm x 23mm x 9mmported in the latest version of Raspbian, Raspberry Pi's preferred operating systemFixed focus lens on-board8 megapixel native resolution sensor-capable of 3280 x 2464 pixel static images
- Weight just over 3gConnects to the Raspberry Pi board via a short ribbon cable (supplied)
- Camera v2 is supported in the latest version of Raspbian, Raspberry Pi's preferred operating system
- megapixel native resolution sensor-capable of 3280 x 2464 pixel static images
- Fixed focus lens on-board
- 8 megapixel native resolution sensor-capable of 3280 x 2464 pixel static images
Apart from these main Boards and Modules, I will be using a soil moisture Sensor and Temperature and Humidity Sensor for Inferencing.
Since the sensors and camera modules for all the plants in an array cumulatively send data to a single Raspberry Pi for processing, Video Processing Units are important to speed up the task as well as reduce the load on Processors. This is the area where Intel Neural Compute Stick 2 comes into picture. This helps in Inferencing, Processing, classification of Video data inputs from over 6 sources in an array at a time which the Raspberry Pi Processor is not capable of. This solution is comparatively quite cost efficient and affordable to be deployed in Apple Farms rather than sending data to the cloud for computation. Sending Video data to the Cloud requires high availability of Internet in remote areas as well as a huge server to store Video data input. Thus resulting in increased Expenses during deploying. Hence, Here I have used a solution which inferences processes data at the Edge.
Training Computer Vision Model for Plant Disease Detection:Automatic and accurate estimation of disease severity is essential for food security, disease management, and yield loss prediction. Deep learning, the latest breakthrough in computer vision, is promising for fine-grained disease severity classification, as the method avoids the labor-intensive feature engineering and threshold-based segmentation. Using the apple disease images in the custom dataset, which are further annotated by botanists with four severity stages as ground truth, a series of deep convolutional neural networks are trained to diagnose the severity of the disease.
Gathering Image Dataset:
Since this model is built to classify Apple Diseases of 6kinds, there was no open Dataset available for all the diseases. Taking this into consideration, I decided to use Google Open Dataset for training the model.
For training the computer Vision Model, I decided to go with YOLOv3 model Architecture.
You only look once, or YOLO, is one of the faster object detection algorithms out there. Though it is no longer the most accurate object detection algorithm, it is a very good choice when you need real-time detection, without loss of too much accuracy.
YOLO v3 uses a variant of Darknet, which originally has 53 layer network trained on Imagenet. For the task of detection, 53 more layers are stacked onto it, giving us a 106 layer fully convolutional underlying architecture for YOLO v3. This is the reason behind the slowness of YOLO v3 compared to YOLO v2. Here is how the architecture of YOLO now looks like.
The most salient feature of v3 is that it makes detections at three different scales. YOLO is a fully convolutional network and its eventual output is generated by applying a 1 x 1 kernel on a feature map. In YOLO v3, the detection is done by applying 1 x 1 detection kernels on feature maps of three different sizes at three different places in the network.The shape of the detection kernel is 1 x 1 x (B x (5 + C) ). Here B is the number of bounding boxes a cell on the feature map can predict, “5” is for the 4 bounding box attributes and one object confidence, and C is the number of classes. In YOLO v3 trained on COCO, B = 3 and C = 80, so the kernel size is 1 x 1 x 255. The feature map produced by this kernel has identical height and width of the previous feature map, and has detection attributes along the depth as described above.
YOLO v3 makes prediction at three scales, which are precisely given by downsampling the dimensions of the input image by 32, 16 and 8 respectively.The first detection is made by the 82nd layer. For the first 81 layers, the image is down sampled by the network, such that the 81st layer has a stride of 32. In this model, we have an image of 416 x 416, the resultant feature map would be of size 13 x 13. One detection is made here using the 1 x 1 detection kernel, giving us a detection feature map of 13 x 13 x 255.Then, the feature map from layer 79 is subjected to a few convolutional layers before being up sampled by 2x to dimensions of 26 x 26. This feature map is then depth concatenated with the feature map from layer 61. Then the combined feature maps is again subjected a few 1 x 1 convolutional layers to fuse the features from the earlier layer (61). Then, the second detection is made by the 94th layer, yielding a detection feature map of 26 x 26 x 255.A similar procedure is followed again, where the feature map from layer 91 is subjected to few convolutional layers before being depth concatenated with a feature map from layer 36. Like before, a few 1 x 1 convolutional layers follow to fuse the information from the previous layer (36). We make the final of the 3 at 106th layer, yielding feature map of size 52 x 52 x 255.
YOLOv3 is Better at detecting smaller objects which is a key feature required to detect diseases in Apple Plant and Leaf🌿Detections at different layers helps address the issue of detecting small objects, a frequent complaint with YOLO v2. The upsampled layers concatenated with the previous layers help preserve the fine grained features which help in detecting small objects.
Using LabelImg to generate Images and Annotations compatible with YOLO format:
LabelImg is a graphical image annotation tool.It is written in Python and uses Qt for its graphical interface.
Annotations are saved as XML files in PASCAL VOC format, the format used by ImageNet. Besides, it also supports YOLO formatAnnotations are saved as XML files in PASCAL VOC format, the format used by ImageNet. Besides, it also supports YOLO format
For this model, I will be using the YOLOv3 format for image Annotation.
In the below Images I have used labelImg to capture data within an image and convert the format to YOLOV3.
Since this dataset needs to be extremely accurate with evasion of background data, use of pre-built Object detection frameworks like Teachable Machine cannot be used for training accurate models which YOLOV3 can perform.
Labelling Images as per YOLOV3 format:
This Image is categorised under "fresh" category which detects fresh shrubs of Apple Plants which are just growing or are still in the process of bearing Apples
These are the Images under the category "ripe". These images are labelled to detect all the Ripe Apples which are ready for harvesting. The main factor of differentiation between the Ripe Apples, Raw Apples and Diseased Apples is the colour. Hence, during training these Images, it is better to use suitable backgrounds representing the colour of the Ripe Apple under the category "ripe". Due to these reasons, the model is trained using RGB and not GrayScale as a basis of differentiation.
These Images are categorised under the category of "Raw Apples". The main factor of differentiation in these Images is the green colour of the Apples. If these Images would have been taken in a leafy background, edge classification of these Images would have not been so accurate. The model would not accurately differentiate between leaves and Green Apples. For this purpose, the images in this category have been labelled in a white background to perform accurate edge detection of Apples. If we take into consideration raw apples in a white background, excluding the Green Nature of the Apples, the complete background is white. In this Image, the colour of the background changes drastically giving the model a factor of differentiation of the Raw Apple. In case of Red/Ripe Apples in a white background, such a basis of differentiation of the object is not required because the colour of the Ripe Apple on the forth is Red while the colour of the background is green, giving the model a basis of discrimination between the background and the Apple.
These Images are categorised under the category "leaf rollers". Leaf rollers are the most likely found pests on an Apple plant as well as source of many diseases for plants. In this category, most of the Images were taken with leaf rollers present on leaves. In this category, the classification is done on the basis of the leaf-roller shape and hence, the background does not contribute to a large extent in decreasing the category of the class. To accurately detect leaf-rollers during implementation of the model, the background was assumed to be the background while carrying out actual inference of the model. For this purpose, the images were taken as representation of actual on-sight leaf-rollers.
The above image is categorised under the class "flowering" which detects the Apple Flowers. This category is used for sending alerts mentioning that since flowers are observed on the plant, It is required to take more care of the plant.
In this category, the main factor of differentiation of the object from other categories is the shape as well as the colour of the flower. The colour of the flower stands to be the major factor of classification in this category. In this category, two types of flowers are taken into consideration which are White flowers(buds of the plants), and the Purple flowers( Fully grown flowers of the plant)
Training the YOLOv3 Model and converting to OpenVino:
Considering the above parameters, and classes and the basis of differentiation of these classes, I decided to go with YOLOv3 framework for object detection.
The YOLOv3 Object detection framework takes into consideration these parameters:
- Bounding Box Prediction:
- tx, ty, tw, th are predicted.
- During training, sum of squared error loss is used.
- And objectness score is predicted using logistic regression. It is 1 if the bounding box prior overlaps a ground truth object by more than any other bounding box prior. Only one bounding box prior is assigned for each ground truth object.
2. Class Prediction
- Softmax is not used.
- Instead, independent logistic classifiers are used and binary cross-entropy loss is used. Because there may be overlapping labels for multilabel classification such as if the YOLOv3 is moved to other more complex domain such as Open Images Dataset.
3. Prediction Across Scales
- 3 different scales are used.
- Features are extracted from these scales like FPN.
- Several convolutional layers are added to the base feature extractor Darknet-53 (which is mentioned in the next section).
- The last of these layers predicts the bounding box, objectness and class predictions.
- On COCO dataset, 3 boxes at each scales. Therefore, the output tensor is N×N×[3×(4+1+80)], i.e. 4 bounding box offsets, 1 objectness prediction, and 80 class predictions.
- Next, the feature map is taken from 2 layers previous and is upsampled by 2×. A feature map is also taken from earlier in the network and merge it with our upsampled features using concatenation. This is actually the typical encoder-decoder architecture, just like SSD is evolved to DSSD.
- This method allows us to get more meaningful semantic information from the upsampled features and finer-grained information from the earlier feature map.
- Then, a few more convolutional layers are added to process this combined feature map, and eventually predict a similar tensor, although now twice the size.
- k-means clustering is used here as well to find better bounding box prior. Finally, on COCO dataset, (10×13), (16×30), (33×23), (30×61), (62×45), (59×119), (116×90), (156×198), and (373×326) are used.
4. Feature Extractor: Darknet-53
Darknet-53
- Darknet-19 classification network is used in YOLOv2 for feature extraction.
- Now, in YOLOv3, a much deeper network Darknet-53 is used, i.e. 53 convolutional layers.
- Both YOLOv2 and YOLOv3 also use Batch Normalization.
- Shortcut connections are also used as shown above.
1000-Class ImageNet Comparison (Bn Ops: Billions of Operations, BFLOP/s: Billion Floating Point Operation Per Second, FPS: Frame Per Second)
- 1000-class ImageNet Top-1 and Top5 error rates are measured as above.
- Single Crop 256×256 image testing is used, on a Titan X GPU.
- Compared with ResNet-101, Darknet-53 has better performance (authors mentioned this in the paper) and it is 1.5× faster.
- Compared with ResNet-152, Darknet-53 has similar performance (authors mentioned this in the paper) and it is 2× faster.
5.1. COCO mAP@0.5
mAP@0.5
- As shown above, compared with RetinaNet, YOLOv3 got comparable mAP@0.5 with much faster inference time.
- For example, YOLOv3–608 got 57.9% mAP in 51ms while RetinaNet-101–800 only got 57.5% mAP in 198ms, which is 3.8× faster.
These parameters mentioned above make YOLOv3 an accurate framework in comparison with RetinaNet - 50 and RetinaNet - 101 and make it significantly faster than these Frameworks. Even after these parameters, which make YOLOv3 easier to deploy on the edge, it is still far heavy to be deployed on Microcontrollers like Raspberry PI. For this purpose, OpenVino is used which quantizes the model further.
Training the YOLOv3 Model:
To train the YOLOv3 model efficiently and decrease the training duration as well as increase the number of training iterations, I decided to train the model on Opensource Google Colab Platform Notebook's.
For the model training process, I have used Google Colab GPU Notebook to increase training efficiency and speed by nearly 100x as compared to a CPU
Cloning AlexeyAB's Darknet Repository:
!git clone https://github.com/AlexeyAB/darknetNote: Syntaxes may be different as compared to terminal because this is in a Jupyter Notebook format
Darknet-53 is a convolutional neural network that acts as a backbone for the YOLOv3 object detection approach. The improvements upon its predecessor Darknet-19 include the use of residual connections, as well as more layers.
Changing Makefile to have GPU and OpenCV enabled:
# change makefile to have GPU and OPENCV enabled
%cd darknet
!sed -i 's/OPENCV=0/OPENCV=1/' Makefile
!sed -i 's/GPU=0/GPU=1/' Makefile
!sed -i 's/CUDNN=0/CUDNN=1/' MakefileThe below code is to verify if the cuda driver is enabled:
# verify CUDA
!/usr/local/cuda/bin/nvcc --versionAfter verifying it, It gives an output as : which shows that the CUDA Driver is enabled on the Colab Notebook:
nvcc: NVIDIA (R) Cuda compiler driver Copyright (c) 2005-2019 NVIDIA Corporation Built on Sun_Jul_28_19:07:16_PDT_2019 Cuda compilation tools, release 10.1, V10.1.243
This function builds the Darknet environment to train the YOLOV3 model:
# make darknet (build)
!makeThe below code defines all the helper functions which are required throughout the training process:
# define helper functions
def imShow(path):
import cv2
import matplotlib.pyplot as plt
%matplotlib inline
image = cv2.imread(path)
height, width = image.shape[:2]
resized_image = cv2.resize(image,(3*width, 3*height), interpolation = cv2.INTER_CUBIC)
fig = plt.gcf()
fig.set_size_inches(18, 10)
plt.axis("off")
plt.imshow(cv2.cvtColor(resized_image, cv2.COLOR_BGR2RGB))
plt.show()
# use this to upload files
def upload():
from google.colab import files
uploaded = files.upload()
for name, data in uploaded.items():
with open(name, 'wb') as f:
f.write(data)
print ('saved file', name)
# use this to download a file
def download(path):
from google.colab import files
files.download(path)Over here, I have defined helper functions like imShow to show output images as well as training graph of val_loss and mAP. Height and Width helper functions are used to set the height and width of input image and resized_image function resizes the images. Besides this, an input file function and file path function has been defined to take file inputs and allow downloading the file path.
Before going ahead with the next steps; the requirements for YOLOv3 need to be downloaded.
After having these files downloaded, we can go ahead and follow the next steps:
In this I am linking the Colab Notebook to Google Drive to transfer the requirements to the Colab directory;
%cd ..
from google.colab import drive
drive.mount('/content/gdrive')
# this creates a symbolic link so that now the path /content/gdrive/My\ Drive/ is equal to /mydrive
!ln -s /content/gdrive/My\ Drive/ /mydrive
!ls /mydriveThis changes the default directory to be the Darknet folder
%cd darknetAfter the environment and variables are set up, I compressed the trained YOLOv3 dataset with images and labels and uploaded it to my drive. The zip folder with Training and Testing dataset is now uploaded to github.
To unzip and copy these images, I used the following:
!ls /mydrive/yolov3
!cp /mydrive/yolov3/obj.zip ../
!unzip ../obj.zip -d data/The cfg file is the most important while training the hydra model.
The important variables of the cfg file were set as follows:
batch = 64
subdivisions = 16
These are recommended integers to attain an accurate model
max_batches = 18000
steps = 14400,16200
These variables vary according to the number of classes in the model.
classes = 9 - [ since the model consists of 9 different classes]
filters = 42
Finally after changing these variables, I uploaded the cfg file to the Colab Notebook to go ahead and train the model:
!cp /mydrive/yolov3/yolov3_custom.cfg ./cfgThe obj.names file consists of all the class names as per YOLO format
In this model, it consists of 9 classes as follows:
- alternaria
- cedar
- fire-blight
- flowering
- fresh
- fungal
- leaf-roller
- raw
- ripe
Out of these 9 classes, 4 are states of the plant and the rest 5 are diseases of plants.
The obj.data file consists of information regarding file path to other files require during the training:
classes = 9
train = data/train.txt
valid = data/test.txt
names = data/obj.names
backup = /mydrive/yolov3/backupAfter configuring these files, I copied both the files to the Colab Notebook:
!cp /mydrive/yolov3/obj.names ./data
!cp /mydrive/yolov3/obj.data ./dataThe next step is to upload image paths to a.txt folder using a python script called generate_train.py
!cp /mydrive/yolov3/generate_train.py ./
!python generate_train.pyThis will generate image paths required in a YOLO format.
After this, I downloaded the weights for the convolutional layers of the YOLOv3 network. By using these weights it helps my object detector to be way more accurate and not have to train as long. Its not necessary to use these weights but it speeds up the process and makes the model accurate.
!wget http://pjreddie.com/media/files/darknet53.conv.74After setting up these requirements, I went ahead to train my model using the following command:
!./darknet detector train data/obj.data cfg/yolov3_custom.cfg darknet53.conv.74 -dont_showThis process took around 6 to 7 hours to complete and completely train the model until the model could be used. The val_loss started from an initial variable of 1230.34 and went till 2.38
using the imshow() function, I checked the graph of the model
This shows the distribution of val_loss across the number of iterations. After training the model to 3500 iterations and reaching a loss of 2.3884, I stopped training the model further and went to check the mAP ( Mean Average Precision ) of the model using the command:
!./darknet detector map data/obj.data cfg/yolov4-obj.cfg /mydrive/yolov3/backup/yolov3_custom2_last.weightsThe mAP of the model was 51.58% is a fairly accurate model. On Average mAP of models do not cross the 58% benchmark in YOLOv3 models.
Some classes performed really well in the model training process while some did not cross the 50% limit. Classes like flowering and Fungal did not perform extremely well in the mAP but during generating the output process, they could predict the classes with a minimum threshold of 0.5
This completes the model training process and to check the model results, I took various images of Apple Plants and some images with diseases to perform inference using the command:
# need to set our custom cfg to test mode
%cd cfg
!sed -i 's/batch=64/batch=1/' yolov3_custom.cfg
!sed -i 's/subdivisions=16/subdivisions=1/' yolov3_custom.cfg
%cd ..
!./darknet detector test data/obj.data cfg/yolov3_custom.cfg /mydrive/yolov3/backup/yolov3_custom_last.weights /mydrive/images/apples.jpg -thresh 0.3
imShow('predictions.jpg')After using this command, I generated output for 6 images which are displayed here:
1) Ripe Apples inference: For this image, it detected all the Apples with an average confidence rating of above 0.9 which indicates the model to be accurate. In this image nearly 13 ripe apples have been detected and a fresh plant in the background is detected which shows a newly growing plant which does not bear fruits or flowers.
2) Leaf Roller detected - The leaf rollers in the image are detected with a confidence score of 0.96. This image displays the plant from a close-up but if the leaf-rollers are located at a distant location, the model detects the leaf-roller with a confidence score of 0.67.
3) Cedar rust detected: In this image, the cedar rust disease is detected with a confidence rating of 0.68. The drop in the confidence score is because of the black background which was not trained in the model. The cedar rust was trained with green natural background and hence on taking an image with a black background, the confidence rating has dropped. On performing the detection with a green background, the confidence increases to 0.86 and 0.91. Thus, this model performs really well in real life environment than demo images.
4) Fire-Blight detected - In this image, fire-blight is detected from a confidence rating of 0.6 to 0.9. All the leaves diagnosed with fire-blight in the image are detected by the Model. Towards the left, the leaf in the pre-stage of fire-blight is detected as well which serves as a warning to the forthcoming diseases.
5) Detection of Raw Apples with Ripe Apples: This image consists of instances with raw apples as well as Ripe apples which is really hard for the model to detect, but this model detects most of the instances and categorises them accordingly. In a few cases, the model classified ripe apples to be raw, but in most of the cases, Apples were detected accurately. The confidence rating of the instances started from 0.4 to 0.9.
5) Detection of Flowering plants: In this image, all the flowers along with buds are detected with a confidence rating of 0.3 to 0.9. Nearly 7 instances of flowers/buds have been detected in the mage which is in indicator that the Apple Plant is doing well. Using these 9 classes of model training, all the conditions of the Apple Plant can be detected from performing Extremely well to performing Critically Bad.
After we have created a YOLOv3 model, it may seem that why is the OpenVino toolkit used to deploy the models on the Raspberry PI:
OpenVINO stands for Open Visual Inference and Neural Network Optimization. It is a toolkit provided by Intel to facilitate faster inference of deep learning models. It helps developers to create cost-effective and robust computer vision applications. It enables deep learning inference at the edge and supports heterogeneous execution across computer vision accelerators — CPU, GPU, Intel® Movidius™ Neural Compute Stick, and FPGA. It supports a large number of deep learning models out of the box.
Architecture of the OpenVino Engine:
The execution process is as follows —
- We feed a pretrained model to the Model Optimizer. It optimizes the model and converts it into its intermediate representation (.xml and.bin file).
- The Inference Engine helps in proper execution of the model on different devices. It manages the libraries required to run the code properly on different platforms.
The two main components of OpenVINO toolkit are Model Optimizer and Inference Engine.
Model Optimizer
Model optimizer is a cross-platform command line tool that facilitates the transition between the training and deployment environment. It adjusts the deep learning models for optimal execution on end-point target devices.
Model Optimizer loads a model into memory, reads it, builds the internal representation of the model, optimizes it, and produces the Intermediate Representation. Intermediate Representation is the only format that the Inference Engine accepts and understands.
The Model Optimizer does not infer models. It is an offline tool that runs before the inference takes place.
The Model Optimizer does not infer models. It is an offline tool that runs before the inference takes place.
Model Optimizer has two main purposes:
- Produce a valid Intermediate Representation. The primary responsibility of the Model Optimizer is to produce two files (.xml and.bin) that forms the Intermediate Representation.
- Produce an optimized Intermediate Representation. Pretrained models contain layers that are important for training, such as the Dropout layer. These layers are useless during inference and might increase the inference time. In many cases, these layers can be automatically removed from the resulting Intermediate Representation. However, if a group of layers can be represented as one mathematical operation, and thus as a single layer, the Model Optimizer recognizes such patterns and replaces these layers with only one. The result is an Intermediate Representation that has fewer layers than the original model. This decreases the inference time.
Operations
- Reshaping —
- The Model Optimizer allows us to reshape our input images. For the Apple disease detection model, I have trained the images with an image size of 412*412 and while deploying the model on the Raspberry Pi, the image size can be changed to 96*96 for faster processing. For this I can simply pass on the new image size as a command line argument and the Model Optimizer will handle the rest.
Batching —
- We can change the batch size of our model at inference time. We can just pass the value of batch size as a command line argument.
- We can also pass our image size like this [4, 3, 100, 100]. Here we are specifying that we need 4 images with dimensions 100*100*3 i.e RGB images having 3 channels and having width and height as 100. Important thing to note here is that now the inference will be slower as we are using a batch of 4 images for inference rather than using just a single image.
Standardizing and Scaling —
- We can perform operations like normalization (mean subtraction) and standardization on our input data.
Quantization
It is an important step in the optimization process. Most deep learning models generally use the FP32 format for their input data. The FP32 format consumes a lot of memory and hence increases the inference time. So, intuitively we may think, that we can reduce our inference time by changing the format of our input data. There are various other formats like FP16 and INT8 which we can use, but we need to be careful while performing quantization as it can also result in loss of accuracy. For deploying the model to the Raspberry Pi, we can quantize the model which leads to an accuracy loss of the model by nearly 5% but decreases the inference time significantly.
Using the INT8 format can help us in reducing our inference time significantly, but currently, only certain layers are compatible with the INT8 format: Convolution, ReLU, Pooling, Eltwise and Concat. So, we essentially perform hybrid execution where some layers use FP32 format whereas some layers use INT8 format. There is a separate layer which handles theses conversions. i.e we don’t have to explicitly specify the type conversion from one layer to another.
Calibrate layer handles all these intricate type conversions. The way it works is as follows —
- Initially, we need to define a threshold value. It determines how much drop in accuracy are we willing to accept.
- The Calibrate layer then takes a subset of data and tries to convert the data format of layers from FP32 to INT8 or FP16.
- It then checks the accuracy drop and if it less than the specified threshold value, then the conversion takes place.
After using the Model Optimizer to create an intermediate representation (IR), we use the Inference Engine to infer input data.
The Inference Engine is a C++ library with a set of C++ classes to infer input data (images) and get a result. The C++ library provides an API to read the Intermediate Representation, set the input and output formats, and execute the model on devices.
The heterogeneous execution of the model is possible because of the Inference Engine. It uses different plug-ins for different devices.
Heterogeneous Plug-in
- We can execute the same program on multiple devices. We just need to pass in the target device as a command line argument and the Inference Engine will take care of the rest i.e. we can run the same piece of code on a CPU, GPU, VPU or any other device compatible with the OpenVINO toolkit. --This is an extremely important feature in the Apple Disease detection model. I will be deploying the model on individual Raspberry Pi microcontrollers corresponding to the array they are deployed to.
- We can also execute parts of our program on different devices i.e. some part of our program might run on CPU and other part might be running on a FPGA or a GPU. If we specify HETERO: FPGA, CPU then the code will run primarily on an FPGA, but if suppose it encounters a particular operation which is not compatible with FPGA then it will switch to CPU.
- We can also execute certain layers on a specific device. Suppose you want to run the Convolution layer only on your GPU then you can explicitly specify it.
Deploying the Model on the Raspberry PI:
The OpenVINO™ toolkit quickly deploys applications and solutions that emulate human vision. Based on Convolutional Neural Networks (CNN), the toolkit extends computer vision (CV) workloads across Intel® hardware, maximizing performance. The OpenVINO toolkit includes the Intel® Deep Learning Deployment Toolkit (Intel® DLDT).
The OpenVINO™ toolkit for Raspbian* OS includes the Inference Engine and the MYRIAD plugins. You can use it with the Intel® Neural Compute Stick 2 plugged in one of USB ports.
Included in the Installation Package
The OpenVINO toolkit for Raspbian OS is an archive with pre-installed header files and libraries. The following components are installed by default:
Hardware
- Raspberry Pi* board with ARM* ARMv7-A CPU architecture.
One of Intel® Movidius™ Visual Processing Units (VPU):
- Intel® Neural Compute Stick 2
- One of Intel® Movidius™ Visual Processing Units (VPU):Intel® Neural Compute Stick 2
Downloading the l_openvino_toolkit_runtime_raspbian_p_2020.4.28 version of the OpenVino Software for Raspberry PI.
- Open the Terminal* or your preferred console application.
- Go to the directory in which you downloaded the OpenVINO toolkit.
cd ~/Downloads/Create an installation folder.
sudo mkdir -p /opt/intel/openvinoUnpack the archive:
sudo tar -xf l_openvino_toolkit_runtime_raspbian_p_2020.4.28.tgz --strip 1 -C /opt/intel/openvinoNow the OpenVINO toolkit components are installed.
Install External Software Dependencies
CMake* version 3.7.2 or higher is required for building the Inference Engine sample application. To install, open a Terminal* window and run the following command:
sudo apt install cmakeSet the Environment Variables
You must update several environment variables before you can compile and run OpenVINO toolkit applications. Run the following script to temporarily set the environment variables:
source /opt/intel/openvino/bin/setupvars.sh**(Optional)** The OpenVINO environment variables are removed when you close the shell. As an option, you can permanently set the environment variables as follows:
echo "source /opt/intel/openvino/bin/setupvars.sh" >> ~/.bashrcTo test your change, open a new terminal. You will see the following:
[setupvars.sh] OpenVINO environment initializedContinue to the next section to add USB rules for Intel® Neural Compute Stick 2 devices.
Add USB Rules
Add the current Linux user to the users group:
sudo usermod -a -G users "$(pi@raspberrypi)"Log out and log in for it to take effect.Add the current Linux user to the users group:
sudo usermod -a -G users "$(pi@raspberrypi)"If you didn't modify .bashrc to permanently set the environment variables, run setupvars.sh again after logging in:
source /opt/intel/openvino/bin/setupvars.shTo perform inference on the Intel® Neural Compute Stick 2, install the USB rules running the install_NCS_udev_rules.sh script:
sh /opt/intel/openvino/install_dependencies/install_NCS_udev_rules- Plug in your Intel® Neural Compute Stick 2.
After the Installation is complete the Raspberry Pi is set up to perform inference.
If you want to use your model for inference, the model must be converted to the.bin and.xml Intermediate Representation (IR) files that are used as input by Inference Engine.
Converting YOLOv3 to OpenVino Intermediate Representation (IR):To deploy the model on the Raspberry PI, we need to YOLOv3* public models to the Intermediate Representation (IR). All YOLO* models are originally implemented in the DarkNet* framework and consist of two files:
.cfgfile with model configurations.weightsfile with model weights
Depending on a YOLO model version, the Model Optimizer converts it differently:
- YOLOv3 has several implementations. This uses a TensorFlow implementation of YOLOv3 model, which can be directly converted to the IR.
Overview of YOLOv3 Model Architecture
Originally, YOLOv3 model includes feature extractor called Darknet-53 with three branches at the end that make detections at three different scales. These branches must end with the YOLO Region layer.
Region layer was first introduced in the DarkNet framework. Other frameworks, including TensorFlow, do not have the Region implemented as a single layer, so every author of public YOLOv3 model creates it using simple layers. This badly affects performance. For this reason, the main idea of YOLOv3 model conversion to IR is to cut off these custom Region-like parts of the model and complete the model with the Region layers where required.
Dump TensorFlow model out of https://github.com/mystic123/tensorflow-yolo-v3 GitHub repository (commit fb9f543)
Clone the repository:
!git clone https://github.com/mystic123/tensorflow-yolo-v3.git
cd tensorflow-yolo-v3Checkout to the commit that the conversion was tested on:
!git checkout fb9f543Run a converter:
--for YOLO-v3:
!python3 convert_weights_pb.py --class_names content/darknet/cfg/coco.names --data_format NHWC --weights_file content/darknet/yolov3.weightsOutput generating a frozen_darknet_yolov3_model.pb file:
/usr/local/lib/python3.6/dist-packages/tensorflow/python/framework/dtypes.py:523: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'. _np_qint8 = np.dtype([("qint8", np.int8, 1)]) /usr/local/lib/python3.6/dist-packages/tensorflow/python/framework/dtypes.py:524: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'. _np_quint8 = np.dtype([("quint8", np.uint8, 1)]) /usr/local/lib/python3.6/dist-packages/tensorflow/python/framework/dtypes.py:525: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'. _np_qint16 = np.dtype([("qint16", np.int16, 1)]) /usr/local/lib/python3.6/dist-packages/tensorflow/python/framework/dtypes.py:526: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'. _np_quint16 = np.dtype([("quint16", np.uint16, 1)]) /usr/local/lib/python3.6/dist-packages/tensorflow/python/framework/dtypes.py:527: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'. _np_qint32 = np.dtype([("qint32", np.int32, 1)]) /usr/local/lib/python3.6/dist-packages/tensorflow/python/framework/dtypes.py:532: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'. np_resource = np.dtype([("resource", np.ubyte, 1)]) 2020-09-26 17:24:24.472975: I tensorflow/core/platform/cpu_feature_guard.cc:141] Your CPU supports instructions that this TensorFlow binary was not compiled to use: AVX2 FMA 1330 ops written to frozen_darknet_yolov3_model.pb.These commands have been deployed on a Google Colab Notebook where the Apple diseases .weights file was located
I have renamed the frozen_darknet_yolov3_model.pb file to yolo_v3.pb
Link to yolo_v3.pb file : YOLOv3 file drive link
This command creates the yolo_v3.pb file, which is a TensorFlow representation of the YOLOv3 model, in the darknet directory.
Convert YOLOv3 TensorFlow Model to the IR
To solve the problems explained in the YOLO V3 architecture overview section, use the yolo_v3.json configuration file with custom operations located in the <OPENVINO_INSTALL_DIR>/deployment_tools/model_optimizer/extensions/front/tf repository.
It consists of several attributes:
[
{
"id": "TFYOLOV3",
"match_kind": "general",
"custom_attributes": {
"classes": 9,
"coords": 4,
"num": 9,
"mask": [0, 1, 2],
"entry_points": ["detector/yolo-v3/Reshape", "detector/yolo-v3/Reshape_4", "detector/yolo-v3/Reshape_8"]
}
}
]where:
idandmatch_kindare parameters that you cannot change.
custom_attributes is a parameter that stores all the YOLOv3 specific attributes:
classes,coords,num, andmaskare attributes that nned to be changed using the configuration file file that was used for model training i.eyolov3_custom2.cfg. Replacing the default values incustom_attributeswith the parameters that follow the[yolo]title in the configuration file. -- here I have set classes =9 and num = 9 with mask = 0, 1, 2custom_attributesis a parameter that stores all the YOLOv3 specific attributes:
To generate the IR of the YOLOv3 TensorFlow model, run:
python3 mo_tf.py
--input_model /path/to/yolo_v3.pb
--tensorflow_use_custom_operations_config $MO_ROOT/extensions/front/tf/yolo_v3.json
--batch 1Optimizing the model:
[ SUCCESS ] Generated IR version 10 model.
[ SUCCESS ] XML file: yolo-openvino-hydra.xml
[ SUCCESS ] BIN file: yolo-openvino-hydra.bin
[ SUCCESS ] Total execution time: 19.16 seconds.After this is created, we get an .xml file to deploy our model on OpenVino:
python3 main.py -d MYRIAD -m yolo-openvino-hydra.xml -pt 0.5After Deploying this command, this activates the camera module deployed on the Raspberry Pi is activated and the inference on the module begins:
This is the timelapse video of a duration of 4 days reduced to 2 seconds. During actual inference of video input, this data is recorded in real time and accordingly real time notifications are updated. These notifications do not change quite frequently because the change in Video data is not a lot.
After I have successfully configured and generated the output video, detection of the video data wont be enough. In that case, I decided to send this video output data to a web-frontend dashboard for other Data-Visualization.
Using this object detection model, I will generate further outputs based on the class_name of the Apple Diseases detected. For this purpose, I have assigned a variable corresponding to the class name = 0: This is a simple logic which generates outputs based on the constant declared. The output generator is as follows:
darknet.copy_image_from_bytes(darknet_image, image_resized.tobytes())
detections = darknet.detect_image(network, class_names, darknet_image, thresh=thresh)
image = darknet.draw_boxes(detections, image_resized, class_colors)
return cv2.cvtColor(image, cv2.COLOR_BGR2RGB), detections
if class_names == "fresh":
fresh = 0
if class_names == "ripe":
ripe = 0
if class_names == "raw":
raw = 0
if class_names == "flowering":
flowering = 0
if class_names == "alternaria":
alternaria = 0
if class_names == "cedar":
cedar = 0
if class_names == "fire-blight":
fire_blight = 0
if class_names == "leaf-roller":
leaf_roller = 0
if class_names == "fungal":
fungal = 0Deploying unoptimised Tensorflow Lite model on Raspberry Pi:
Tensorflow Lite is an open-source framework created to run Tensorflow models on mobile devices, IoT devices, and embedded devices. It optimizes the model so that it uses a very low amount of resources from your phone or edge devices like Raspberry Pi
Unlike x86-based systems, most ARM systems (like those on CSP) do not have a supported pre-built TensorFlow package (i.e. installation isn’t as simple as pip install tensorflow). Furthermore, on embedded systems with limited memory and compute, the Python frontend adds substantial overhead to the system and makes inference slow.
Before the release of the TensorFlow Lite developer preview in November 2017, we were exploring the use of the full TensorFlow C++ library. To determine the feasibility of CNNs on embedded hardware we trained a simple LeNet-5 model for the MNIST dataset and benchmarked the performance of the TensorFlow C++ library on CSP. After the release of TensorFlow Lite, we benchmarked the performance of TensorFlow Lite on the same MNIST example.
The main drawback of vanilla TensorFlow C++ on CSP was high memory usage. TensorFlow Lite provides faster execution and lower memory usage compared to vanilla TensorFlow.
By default, Tensorflow Lite interprets a model once it is in a Flatbuffer file format (.tflite) which is generated by the Tensorflow Converter. Before this can be done, we need to convert the darknet model to the Tensorflow supported Protobuf file format (.pb)
I have already converted the file in the above conversion and the link to the pb file is: YOLOv3 file
I used mystic123’s implementation of Yolo v3 in TF-Slim to do the conversions. Before cloning this repository, you need to have Python version ≥ 3.5, and Tensorflow version ≥ 1.11.
.Pb -> .Tflite conversion
To perform this conversion, you need to identify the name of the input, dimensions of the input, and the name of the output of the model.
To run the command below you will need tflite_convert to be installed.
tflite_convert --graph_def_file=yolo_v3.pb --output_file=yolov3-tiny.tflite --input_format=TENSORFLOW_GRAPHDEF --output_format=TFLITE --input_shape=1,416,416,3 --input_array=~YOUR INPUT NAME~ --output_array=~YOUR OUTPUT NAME~ --inference_type=FLOAT --input_data_type=FLOATThis generates a file called yolov3-tiny.tflite
Running TFlite on Raspberry Pi
Let's make sure the camera interface is enabled in the Raspberry Pi Configuration menu. Click the Pi icon in the top left corner of the screen, select Preferences -> Raspberry Pi Configuration, and go to the Interfaces tab and verify Camera is set to Enabled.
Install virtualenv by issuing:
sudo pip3 install virtualenvThen, create the "tflite1-env" virtual environment by issuing:
python3 -m venv tflite1-envThis will create a folder called tflite1-env inside the tflite1 directory. The tflite1-env folder will hold all the package libraries for this environment. Next, activate the environment by issuing:
source tflite1-env/bin/activateYou'll need to issue the source tflite1-env/bin/activate command from inside the /home/pi/tflite1 directory to reactivate the environment every time you open a new terminal window. You can tell when the environment is active by checking if (tflite1-env) appears before the path in your command prompt, as shown in the screenshot below.
At this point, here's what your tflite1 directory should look like if you issue ls.
Step 1c. Install TensorFlow Lite dependencies and OpenCV
Next, we'll install TensorFlow, OpenCV, and all the dependencies needed for both packages. OpenCV is not needed to run TensorFlow Lite, but the object detection scripts in this repository use it to grab images and draw detection results on them.
Clone Repository:
git clone https://github.com/EdjeElectronics/TensorFlow-Lite-Object-Detection-on-Android-and-Raspberry-Pi.gitNext, we'll install TensorFlow, OpenCV, and all the dependencies needed for both packages. OpenCV is not needed to run TensorFlow Lite, but the object detection scripts in this repository use it to grab images and draw detection results on them.
Initiate a shell script that will automatically download and install all the packages and dependencies. Run it by issuing:
bash get_pi_requirements.shStep 1d. Set up TensorFlow Lite detection model
Next, we'll set up the detection model that will be used with TensorFlow Lite.
Add the yolo_v3.tflite model and classes.txt to yolo_v3 folder to run the model
Run the TensorFlow Lite model!
Run the real-time webcam detection script by issuing the following command from inside the /home/pi/tflite1 directory. (Before running the command, make sure the tflite1-env environment is active by checking that (tflite1-env) appears in front of the command prompt.)
python3 TFLite_detection_webcam.py --modeldir=yolo_v3Unoptimized model decrease in accuracy and FPS:
Getting Inferencing results and comparing them:
These are the inferencing results of deploying tensorflow and tflite to Raspberry Pi respectively. Even though the inferencing time in tflite model is less than tensorflow, it is comparitively high to be deployed.
After Using NCS2 on Raspberry Pi :
While deploying the unoptimised model on Raspberry Pi, the CPU Temperature rises drastically and results in poor execution of the model:
Last time the Raspberry Pi reached a temperature of 74°C during extended testing which meant that it suffered from thermal throttling of the CPU, it came close to the 80°C point where additional incremental throttling would occur. This time we saw increased temperatures, peaking around 78°C.
Tensorflow Lite uses 15Mb of memory and this usage peaks to 45mb when the temperature of the CPU rises after performing continuous execution:
Power Consumption while performing inference: In order to reduce the impact of the operating system on the performance, the booting process of the RPi does not start needless processes and services that could cause the processor to waste power and clock cycles in other tasks. Under these conditions, when idle, the system consumes around 1.3 to 1.4 watts
Comparing Frames per Second FPS of the model without NCS2 and with NCS2:
This shows significant jump from 0.63FPS to 8.31FPS Respectively. This increases the model performance by a significant amount which is nearly 12 times.
This increment in FPS and model inferencing is useful when deploying the model on drones using hyperspectral Imaging
Temperature Difference in 2 scenarios in deploying the model:
1) Deploying tensorflow lite unoptimised model on Raspberry Pi without NCS2:
This image shows that the temperature of the core microprocessor rises to a tremendous extent. This is the prediction of the scenario while the model completed 21 seconds after being deployed on the Raspberry Pi. After 121seconds of running the inference, the model crashed and the model had to be restarted again after 4mins of being idle.
2) Deploying OpenVino optimised model with NCS2 on Raspberry Pi:
This image was taken after disconnecting power peripherals and NCS2 from the Raspberry Pi 6 seconds after inferencing. The model ran for about 241seconds without any interruption after which the peripherals were disconnected and the thermal image was taken. This shows that the OpenVino model performs way better than the unoptimised tensorflow lite model and runs smoother. Its also observed that the accuracy of the model increases if the model runs smoothly.
Integrating Additional Sensors with Raspberry PI for complete analysis:1) Integrating Soil Moisture Sensor with the Raspberry Pi:
This is Soil Moisture Meter, Soil Humidity Sensor, Water Sensor, Soil Hygrometer. With this module, you can tell when your plants need watering by how moist the soil is in your pot, garden, or yard. The two probes on the sensor act as variable resistors. Use it in a home automated watering system, hook it up to IoT, or just use it to find out when your plant needs a little love. Installing this sensor and its PCB will have you on your way to growing a green thumb!
The soil moisture sensor consists of two probes which are used to measure the volumetric content of water. The two probes allow the current to pass through the soil and then it gets the resistance value to measure the moisture value.
When there is more water, the soil will conduct more electricity which means that there will be less resistance. Therefore, the moisture level will be higher. Dry soil conducts electricity poorly, so when there will be less water, then the soil will conduct less electricity which means that there will be more resistance. Therefore, the moisture level will be lower.
The sensor board itself has both analogue and digital outputs. The Analogue output gives a variable voltage reading that allows you to estimate the moisture content of the soil. The digital output gives you a simple "on" or "off" when the soil moisture content is above a certain threshold. The value can be set or calibrated using an adjustable on board potentiometer. In this case, we just want to know either "Yes, the plant has enough water" or "No, the plant needs watering!", so we'll be using the digital out. We'll set this value later at which you want the sensor to "trigger" a notification.
Wiring the sensor to the Raspberry Pi.
VCC --> 3v3 (Pin 1)GND --> GND (Pin 9)D0 --> GPIO 17 (Pin 11)
With everything now wired up, we can turn on the Raspberry Pi. Without writing any code we can test to see our moisture sensor working. When power is applied you should see the power light illuminate (with the 4 pins facing down, the power led is the one on the right).
When the sensor detects moisture, a second led will illuminate (with the 4 pins facing down, the moisture detected led is on the left).
I adjusted the potentiometer on the sensor which allows to change the detection threshold (this only applies to the digital output signal) --changing the detection threshold after inferencing some observations, I changed the moisture level threshold to 80%
Now we can see the sensor working, In this model, I want to monitor the moisture levels of the plant pot. So I set the detection point at a level so that if it drops below we get notified that our plant pot is too dry and needs watering. Our plant here, is a little on the dry side, but ok for now, if it gets any drier it’ll need watering. After the moisture sensor is set up to take readings and inference outputs, I will add a peristaltic pump using a relay to perform autonomous Plant Watering.
I’m going to adjust the potentiometer to a point where the detection led just illuminates. As soon as I reach the detection point, I’ll stop turning the potentiometer. That way, when then moisture levels reduce just a small amount the detection led will go out.
The way the digital output works is, when the sensor detects moisture, the output is LOW (0V). When the sensor can no longer detect moisture the output is HIGH (3.3V)
Schematic and Circuit Diagram:
Water Sensor - plug the positive lead from the water sensor to pin 2, and the negative lead to pin 6. Plug the signal wire (yellow) to pin 8.
Relay - Plug the positive lead from pin 7 to IN1 on the Relay Board. Also connect Pin 2 to VCC, and Pin 5 to GND on the Relay board.
Pump - Connect your pump to a power source, run the black ground wire between slots B and C of relay module 1 (when the RPi sends a LOW signal of 0v to pin 1, this will close the circuit turning on the pump).
GPIO Scriptfor Soil Moisture Sensor:
import RPi.GPIO as GPIO
import datetime
import time
init = False
GPIO.setmode(GPIO.BOARD) # Broadcom pin-numbering scheme
def get_last_watered():
try:
f = open("last_watered.txt", "r")
return f.readline()
except:
return "NEVER!"
def get_status(pin = 8):
GPIO.setup(pin, GPIO.IN)
return GPIO.input(pin)
def init_output(pin):
GPIO.setup(pin, GPIO.OUT)
GPIO.output(pin, GPIO.LOW)
GPIO.output(pin, GPIO.HIGH)
def auto_water(delay = 5, pump_pin = 7, water_sensor_pin = 8):
consecutive_water_count = 0
init_output(pump_pin)
print("Here we go! Press CTRL+C to exit")
try:
while 1 and consecutive_water_count < 10:
time.sleep(delay)
wet = get_status(pin = water_sensor_pin) == 0
if not wet:
if consecutive_water_count < 5:
pump_on(pump_pin, 1)
consecutive_water_count += 1
else:
consecutive_water_count = 0
except KeyboardInterrupt: # If CTRL+C is pressed, exit cleanly:
GPIO.cleanup() # cleanup all GPI
def pump_on(pump_pin = 7, delay = 1):
init_output(pump_pin)
f = open("last_watered.txt", "w")
f.write("Last watered {}".format(datetime.datetime.now()))
f.close()
GPIO.output(pump_pin, GPIO.LOW)
time.sleep(1)
GPIO.output(pump_pin, GPIO.HIGH)In the above code snippet, pump in has been set to pin7 and Soil Moisture Sensor pin has been set to pin8.
Over here, a state of the soil moisture sensor has been set to Wet which is a variable continuously aggregating Sensor data. If the Sensor is not found to be wet and if the moisture is below the certain threshold set on the module, it activates the peristaltic pump to start watering the Apple Plant. Similarly, each time it waters the plant, the time when the peristaltic pump was switched on, is printed to last_watered.txt file.
The state of the moisture sensor, If wet or not wet at a particular time is projected on a Streamlit front-end dashboard for Data Visualization. This Front-end data will be displayed in the further part of the project.
2) Integrating Temperature and Humidity Sensor with the Raspberry Pi:
DHT11 is a Digital Sensor consisting of two different sensors in a single package. The sensor contains an NTC (Negative Temperature Coefficient) Temperature Sensor, a Resistive-type Humidity Sensor and an 8-bit Microcontroller to convert the analog signals from these sensors and produce a Digital Output.
DHT11 uses a Single bus data format for communication. Only a single data line between an MCU like Raspberry Pi and the DHT11 Sensor is sufficient for exchange of information.
In this setup, the Microcontroller acts as a Master and the DHT11 Sensor acts as a Slave. The Data OUT of the DHT11 Sensor is in open-drain configuration and hence it must always be pulled HIGH with the help of a 5.1KΩ Resistor.
This pull-up will ensure that the status of the Data is HIGH when the Master doesn’t request the data (DHT11 will not send the data unless requested by the Master).
Now, we will the how the data is transmitted and the data format of the DHT11 Sensor. Whenever the Microcontroller wants to acquire information from DHT11 Sensor
Microcontroller is configured as OUTPUT and it will make the Data Line low for a minimum time of 18ms and releases the line. After this, the Microcontroller pin is made as INPUT.
The data pin of the DHT11 Sensor, which is an INPUT pin, reads the LOW made by the Microcontroller and acts as an OUTPUT pin and sends a response of LOW signal on the data line for about 80µs and then pulls-up the line for another 80µs.
After this, the DHT11 Sensor sends a 40 bit data with Logic ‘0’ being a combination of 50µs of LOW and 26 to 28µs of HIGH and Logic ‘1’ being 50µs of LOW and 70 to 80µs of HIGH.
After transmitting 40 bits of data, the DHT11 Data Pin stays LOW for another 50µs and finally changes its state to input to accept the request from the Microcontroller.
Since we are using a library called Adafruit_DHT provided by Adafruit for this project, we need to first install this library into Raspberry Pi.
git clone https://github.com/adafruit/Adafruit_Python_DHT.gitGPIO Script for Temperature and Humidity Sensor:
import sys
import Adafruit_DHT
import time
while True:
humidity, temperature = Adafruit_DHT.read_retry(11, 4)
print('Temp: {0:0.1f} C Humidity: {1:0.1f} %'.format(temperature, humidity))
time.sleep(1)
if temperature < 8:
success = 0
if (temperature > 8 and temperature < 15):
success = 1
if (temperature > 15 and temperature < 21):
warning = 0
if temperature > 21:
error = 0
if humidity < 70:
success1 = 0
if (humidity > 70 and humidity < 75):
success1 = 1
if (temperature > 75 and temperature < 85):
warning1 = 0
if temperature > 85:
error1 = 0
#this is the data for a single plant in array1 - This code follows the same with other plants changing the coefficient of the error/warning/success outputOn detection of temperature above certain threshold or below certain threshold, variables are assigned with a constant value. If the temperature is below 8C, a variable called success = 0 is defined and this variable is taken into consideration while plotting this data on the Streamlit dashboard. Same goes with humidity sensor. If the humidity is below a certain threshold of 70%, a variable called success1 is assigned a constant called 0.
Configuring Data sorting according to DateTime:
import datetime
now = datetime.datetime.now()
print ("Current date and time : ")
#print (now.strftime("%Y-%m-%d %H:%M:%S"))
x = int(now.strftime("%H"))
print(x)
if x == 0:
array1t0 == int(temperature)
array1h0 == int(humidity)
if x == 1:
array1t1 == int(temperature)
array1h1 == int(humidity)
if x == 2:
array1t2 == int(temperature)
array1h2 == int(humidity)
if x == 3:
array1t3 == int(temperature)
array1h3 == int(humidity)
if x == 4:
array1t4 == int(temperature)
array1h4 == int(humidity)
if x == 5:
array1t5 == int(temperature)
array1h5 == int(humidity)
if x == 6:
array1t6 == int(temperature)
array1h6 == int(humidity)
if x == 7:
array1t7 == int(temperature)
array1h7 == int(humidity)
if x == 8:
array1t8 == int(temperature)
array1h8 == int(humidity)
if x == 9:
array1t9 == int(temperature)
array1h9 == int(humidity)
if x == 10:
array1t10 == int(temperature)
array1h10 == int(humidity)
if x == 11:
array1t11 == int(temperature)
array1h11 == int(humidity)
if x == 12:
array1t12 == int(temperature)
array1h112 == int(humidity)
if x == 13:
array1t13 == int(temperature)
array1h13 == int(humidity)
if x == 14:
array1t14 == int(temperature)
array1h14 == int(humidity)
if x == 15:
array1t15 == int(temperature)
array1h15 == int(humidity)
if x == 16:
array1t16 == int(temperature)
array1h16 == int(humidity)
if x == 17:
array1t17 == int(temperature)
array1h17 == int(humidity)
if x == 18:
array1t18 == int(temperature)
array1h18 == int(humidity)
if x == 19:
array1t19 == int(temperature)
array1h119 == int(humidity)
if x == 20:
array1t20 == int(temperature)
array1h20 == int(humidity)
if x == 21:
array1t21 == int(temperature)
array1h21 == int(humidity)
if x == 22:
array1t22 == int(temperature)
array1h22 == int(humidity)
if x == 23:
array1t23 == int(temperature)
array1h23 == int(humidity)
if x == 24:
array1t24 == int(temperature)
array1h24 == int(humidity)In this script, I have imported DateTime to assign temperature and Humidity sensor data with a timestamp. This is required for Visualisation of Timely Trends in Data. From DateTime I have taken into consideration allocation of Hourly timestamps as per data. Every hour, the temperature data changes and these variables are further utilized for data plotting in Streamlit.
The below video shows the Back-end of the complete project in action:
Functioning of Sensor Logic Algorithms:The soil moisture sensor as well as the humidity and temperature sensor send data readings with assigned timestamps to Network Gateways. These Gateways take this data, sort the data, perform computation and send this data to web cloud application. Here, the Network Gateways are the Raspberry Pi devices. The camera module takes in video data and sends it to the Raspberry PI for classification. The Raspberry PI processes the data and using the Intel Neural Stick 2 and the OpenVino framework, the data is processed and VPU's classify video data. This data is assigned timestamp and further, this classified data is sent to the Streamlit Web Application Front-end Cloud Server. Using Kepler Geo-spatial analysis with satellite Imaging, this data is plotted on a Kpler map for data visualisation with Timely Trends of data. This data is then made availabel after processing to Mobile Users of the farm to analyse the farm and Apple Plantation data, diseases of plant.
Configuring Streamlit Front-end:Streamlit is an awesome new tool that allows engineers to quickly build highly interactive web applications around their data, machine learning models, and pretty much anything.
Installing Streamlit :
pip install streamlitThe below is the code for the streamlit Front-end App.
import streamlit as st
import plotly.graph_objects as go
from PIL import Image
PAGE_CONFIG = {"page_title":"Hydra-OpenVino_Tool","page_icon":":bar_chart:","layout":"centered"}
st.beta_set_page_config(**PAGE_CONFIG)
def main():
st.title("Hydra: OpenVino & AI based Remote plant monitoring & Feeding")
st.subheader("Hydra is an Intel OpenVino based Apple Vegetation Monitoring and Watering system with Visual and Geospatial Analysis based on AIoT Technology")
if __name__ == '__main__':
main()
#Right now, we have used demo data here due to insufficient data from a huge amount of sensors
fig = go.Figure(data=[go.Scatter(
x=[2, 2, 2, 2, 2, 2, 2, 2, 2 ,2 ,2 ,2 ,2 ,2,
4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4,
6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6,
8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8,
10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10,
12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12],
y=[100, 250, 345, 450, 560, 680, 790, 910, 1050, 1250, 1380, 1490, 1670, 1789,
100, 250, 345, 450, 560, 680, 790, 910, 1050, 1250, 1380, 1490, 1670, 1789,
100, 250, 345, 450, 560, 680, 790, 910, 1050, 1250, 1380, 1490, 1670, 1789,
100, 250, 345, 450, 560, 680, 790, 910, 1050, 1250, 1380, 1490, 1670, 1789,
100, 250, 345, 450, 560, 680, 790, 910, 1050, 1250, 1380, 1490, 1670, 1789,
100, 250, 345, 450, 560, 680, 790, 910, 1050, 1250, 1380, 1490, 1670, 1789,],
mode='markers',
marker=dict(
color=[80, 80, 80, 80, 90, 80, 80 , 90, 85, 90, 95, 80, 70, 75,
80, 80, 80, 80, 85, 90, 95, 80, 70, 75, 90, 80, 80 , 90,
70, 75, 80, 90, 70, 75, 80 , 95, 90, 95, 85, 80, 75, 80,
80, 90, 80, 80, 90, 80, 80, 80, 85, 90, 95, 80, 70, 75,
80, 80, 70, 75, 80, 80, 80, 90, 80, 80 , 90, 85, 90, 95,
90, 95, 80, 70, 75, 80, 80, 80, 80, 90, 80, 80 , 90, 85],
size=[25, 25, 25, 25, 30, 25, 25, 30, 28, 30, 35, 25, 20, 23,
25, 25, 25, 25, 30, 28, 30, 35, 25, 20, 23, 30, 25, 25,
20, 23, 25, 30, 20, 23, 25, 35, 30, 35, 28, 25, 23, 25,
25, 30, 25, 25, 25, 25, 25, 30, 28, 30, 35, 25, 20, 23,
25, 25, 20, 23, 25, 25, 25, 30, 25, 25, 30, 28, 30, 35,
30, 35, 25, 20, 23, 25, 25, 25, 25, 30, 25, 25, 30, 28],
showscale=True
)
)])
# 70 - 20,
# 75 - 23,
#80 - 25,
#85 - 28,
#90 - 30,
#95 - 35
fig2 = go.Figure(
data=[go.Bar(y=[array1t6, array1t7, array1t8, array1t9, array1t10, array1t11, array1t12, array1t13, array1t14, array1t15, array1t16, array1t17,
array1t18, array1t19, array1t20, array1t21, array1t22, array1t23, array1t24, array1t0, array1t1, array1t2, array1t3, array1t4, array1t5])],
layout_title_text="Average Temperature of all the Arrays starting from 6am ending at 6am the next day"
)
fig3 = go.Figure(
data=[go.Bar(y=[array1h6, array1h7, array1h8, array1h9, array1h10, array1h11, array1h12, array1h13, array1h14, array1h15, array1h16, array1h17,
array1h18, array1h19, array1h20, array1h21, array1h22, array1h23, array1h24, array1h0, array1h1, array1h2, array1h3, array1h4, array1h5])],
layout_title_text="Average Humididty of all the Arrays starting from 6am ending at 6am the next day"
)
menu = ["Home","Average-Temp","Average-Humidity","Average-Moisture","Geospatial-analysis"]
choice = st.sidebar.selectbox('Menu',menu)
img = Image.open("/content/top-down-farm.jpg")
img2 = Image.open("/content/agri-farm.png")
img3 = Image.open("/content/keplergl.jpeg")
st.sidebar.subheader("Farm plot")
st.sidebar.image(img3, height=250, width=250)
st.sidebar.image(img, height=250, width=250)
st.sidebar.image(img2, height=250, width=250)
if choice == 'Average-Temp':
st.plotly_chart(fig2)
st.write("................................6am yesterday............................................6am today...................................")
if choice == 'Average-Moisture':
st.subheader("This shows the visualisation of the Apple farm starting from Array1 and ending at Array6")
st.header("Push the below button to activate the watering pump in "n1)
if st.button("Activate Pump"):
st.write("Watering pump activated. Plants are now being watered!")
GPIO.output(pump_pin, GPIO.HIGH)
st.header("Push the below button to deactivate the watering pump in "n1)
if st.button("Deactivate Pump"):
st.write("Watering pump deactivated")
GPIO.output(pump_pin, GPIO.LOW)
st.plotly_chart(fig)
if choice == 'Average-Humidity':
st.plotly_chart(fig3)
if choice == 'Geospatial-analysis':
st.image(img3, height=1000, width=1000)
import webbrowser
url = 'https://kepler.gl/demo/map/carto?mapId=2fd6c7b7-6c63-7397-36dc-96a18bd1b900&owner=dhruvsheth-ai&privateMap=false'
if st.button('Open browser'):
webbrowser.open_new_tab(url)
st.write("https://kepler.gl/demo/map/carto?mapId=2fd6c7b7-6c63-7397-36dc-96a18bd1b900&owner=dhruvsheth-ai&privateMap=false")
#average-temperature data to push notifications:
avg_temp = ((array1t6+array1t7+array1t8+array1t9+array1t10+array1t11+array1t12+array1t13+array1t14+array1t15+array1t16+array1t17+array1t18+array1t19+array1t20+array1t21+array1t22+array1t23+array1t24+array1t0+array1t1+array1t2+array1t3+array1t4+array1t5)/24)
avg_hum = ((array1h6+array1h7+array1h8+array1h9+array1h10+array1h11+array1h12+array1h13+array1h14+array1h15+array1h16+array1h17+array1h18+array1h19+array1h20+array1h21+array1h22+array1h23+array1h24+array1h0+array1h1+array1h2+array1h3+array1h4+array1h5)/24)
if choice == "Home":
n1 = "array1"
if st.checkbox("Show Notifications for Diseases"):
if alternaria = 0:
st.error("Alternaria detected in"n1)
elif fresh = 0:
st.success("Fresh trees in "n1)
elif ripe = 0:
st.success("Fresh Apples in "n1)
elif raw = 0:
st.warning("Raw Apples in "n1)
elif cedar = 0:
st.error("Cedar detected in "n1)
elif flowering = 0:
st.success("Flowers in "n1)
elif fungal = 0:
st.error("Fungal Leaves detected in "n1)
elif leaf_roller = 0:
st.error("Leaf Rollers and pests detected in "n1)
elif fire_blight = 0:
st.error("Fire Blight in "n1)
if st.checkbox("Show Notifications for Temperature"):
st.subheader("Normal Temperature = 8C")
st.subheader("Above Average Threshold = 15C")
st.subheader("Critically High Temperature = 20C")
if int(avg_temp) < 8:
st.success("Temperature conducive - lower than 8C in "n1)
elif int(avg_temp) > 8:
st.success("Temperature conducive - higher than 8C in "n1)
elif int(avg_temp) > 15:
st.warning("Temperature above normal - higher than 15C in "n1)
elif int(avg_temp) > 20:
st.error("Temperature critically high - above 20C in "n1)
if st.checkbox("Show notifications for Atmosphere Humidity"):
st.subheader("Normal Humidity = 75%")
st.subheader("Above Average Threshold = 80%")
st.subheader("Critically High Humidity = 85%")
if int(avg_hum) < 75:
st.warning("humidity below normal - lower than 75% in "n1)
elif int(avg_hum) > 75:
st.success("humidity conducive - higher than 75% in "n1)
elif int(avg_hum) > 80:
st.warning("humidity above normal - higher than 80% in "n1)
elif int(avg_hum) > 85:
st.error("humidity critically high - above 85% in "n1)
if st.checkbox("Show notifications for Soil Moisture"):
if wet:
st.warning("Soil moisture levels above 80% in "n1)
if not wet:
#wet variable defined in first cell
st.success("Soil moisture levels below 80% in "n1)
st.subheader("To manually activate pump, locate to Average-Moisture")Understanding the front-end Streamlit code:
Over here, to plot data of soil-moisture of 6 arrays, with nearly 6 plants in each array, we need nearly 36 sensors deployed to produce the inference. Since, these many sensors were not available for the prototype, I have created demo data of Soil Moisture to visualize the data over the plot of land. In case of pushing the notifications on the homescreen, I have taken in data from the Soil Moisture Sensor and this displays whether the plant has moisture above 80% or below 80%. Alternatively, the streamlit dashboard supports manual pump activation to activate the peristaltic pump and water the plants. Usually, the plant is autonomously watered based on water moisture in the soil, but in case if there is manual assistance needed, this trigger allows to activate the pump. The logic used over here is that, each time a button is pressed to activate or deactivate the pump, the GPIO pin is either set to high or low as follows:
if st.button("Activate Pump"):
st.write("Watering pump activated. Plants are now being watered!")
GPIO.output(pump_pin, GPIO.HIGH)
st.header("Push the below button to deactivate the watering pump in "n1)
if st.button("Deactivate Pump"):
st.write("Watering pump deactivated")
GPIO.output(pump_pin, GPIO.LOW)The second figure is meant to display the Temperature data over time. In the above code snippets, I had assigned each hourly sensor data a timestamp. This sensor data with timestamp is taken and added to the plotly chart for visualisation of data with time from 6am in the morning to 6 am the next day. For visualization of this data, the respective data timestamp is assigned with the hour of the day to sync data. This complete process is autonomous. Finally, an average variable for temperature is declared for all the variables over time and this average variable is used to trigger notifications on the notification page as follows:
if int(avg_temp) < 8:
st.success("Temperature conducive - lower than 8C in "n1)
elif int(avg_temp) > 8:
st.success("Temperature conducive - higher than 8C in "n1)
elif int(avg_temp) > 15:
st.warning("Temperature above normal - higher than 15C in "n1)
elif int(avg_temp) > 20:
st.error("Temperature critically high - above 20C in "n1)The third figure is mean to display timely-trend of humidity over time. The process of aggregating and displaying humidity data is the same as tempeerature data. In the above code snippets, I had assigned each hourly sensor data a timestamp. This sensor data with timestamp is taken and added to the plotly chart for visualisation of data with time from 6am in the morning to 6 am the next day. For visualization of this data, the respective data timestamp is assigned with the hour of the day to sync data. This complete process is autonomous. Finally, an average variable for humidity is declared for all the variables over time and this average variable is used to trigger notifications on the notification page as follows:
if int(avg_hum) < 75:
st.warning("humidity below normal - lower than 75% in "n1)
elif int(avg_hum) > 75:
st.success("humidity conducive - higher than 75% in "n1)
elif int(avg_hum) > 80:
st.warning("humidity above normal - higher than 80% in "n1)
elif int(avg_hum) > 85:
st.error("humidity critically high - above 85% in "n1)The fourth figure is meant to display the plot for cumulative diseases detected in a particular array. In the above Object detection toolkit, I have altered the darknet video and image analysis python file to give output each time a particular class name is detected. On detection of a specific disease in aplant for eg- alternaria, a variable called "alternaria" is assigned = 0. In the streamlit front-end code, each time the variable is detected to be 0, the pie chart is updated increasing the percentage share of the disease in the pie chart.
The Notifications page is used for triggering notifications and updates on the health of the plant based on the OpenVino model data input deployed on the Raspberry Pi. The notifications page displays diseases updates over time as follows -- based on the code snippet:
if st.checkbox("Show Notifications for Diseases"):
if alternaria = 0:
st.error("Alternaria detected in"n1)
elif fresh = 0:
st.success("Fresh trees in "n1)
elif ripe = 0:
st.success("Fresh Apples in "n1)
elif raw = 0:
st.warning("Raw Apples in "n1)
elif cedar = 0:
st.error("Cedar detected in "n1)
elif flowering = 0:
st.success("Flowers in "n1)
elif fungal = 0:
st.error("Fungal Leaves detected in "n1)
elif leaf_roller = 0:
st.error("Leaf Rollers and pests detected in "n1)
elif fire_blight = 0:
st.error("Fire Blight in "n1)All these variables were declared in the Darknet script edited earlier in the Object Detection part, so whenever, a class is detected, it assigns the constant value of 0 to the respective class name.
This shows the alerts generated when a disease is detected and a greenpopup box when a ripe apple of a flowering plant is detected.
The home page also displays notifications regarding Temperature, humidity and Soil Moisture Data over time as follows:
The last page is dedicated for Geo-spatial Analysis of data using satellite imaging and data plotting over satellite maps corresponding to the latitude and longitude location and plant plot. For this geo-spatial analysis plot, I have used Kepler.gl, a powerful geo-spatial analysis tool for geo data plotting over time. The streamlit dashboard links the web page to the Kepler.gl tool for futhere analysis:
Video demonstration of Streamlit web app:
Link to the streamlit web app: streamlit-hydra-frontend
Using Kepler.gl tool for Geo-Spatial Data Analysis:Kepler.gl is an advanced geospatial visualization tool open sourced by Uber’s Visualization team in 2018 and contributed to the Urban Computing Foundation earlier this year. At Uber, kepler.gl is the defacto tool for geospatial data analysis.
In order to help data scientists work more effectively, we integrated kepler.gl into many widely used data analysis platforms, now including Jupyter Notebook. Jupyter Notebook is a popular open source web application used to create and share documents that contain live code, equations, visualizations, and text, commonly used among data scientists to conduct data analysis and share results. At Uber, data scientists have utilized this integration to analyze multitudes of geospatial data collected through the app, in order to better understand how people use Uber, and how to improve their trip experience. Now, everyone can leverage kepler.gl within Jupyter Notebook.
Kepler Geo-spatial tool works based on data input from csv, so to configure temperature, humidity and moisture data over time, I will use the pd.write() function to write data to csv file.
#sending data to csv:
import pandas as pd
csv = {'Temperature': ['array1t6', 'array1t7', 'array1t8', 'array1t9', 'array1t10', 'array1t11', 'array1t12', 'array1t13', 'array1t14', 'array1t15', 'array1t16', 'array1t17',
'array1t18', 'array1t19', 'array1t20', 'array1t21', 'array1t22', 'array1t23', 'array1t24', 'array1t0', 'array1t1', 'array1t2', 'array1t3', 'array1t4', 'array1t5'],
'Humidity': ['array1h6', 'array1h7', 'array1h8', 'array1h9', 'array1h10', 'array1h11', 'array1h12', 'array1h13', 'array1h14', 'array1h15', 'array1h16', 'array1h17',
'array1h18', 'array1h19', 'array1h20', 'array1h21', 'array1h22', 'array1h23', 'array1h24', 'array1h0', 'array1h1', 'array1h2', 'array1h3', 'array1h4', 'array1h5'],
'Longitude': [-138, -138, -138, -138, -138, -138, -138, -138, -138, -138, -138, -138, -138, -138, -138, -138, -138, -138, -138, -138, -138, -138, -138, -138],
'Lattitude': [32, 32, 32, 32, 32, 32, 32, 32, 32, 32, 32, 32, 32, 32, 32, 32, 32, 32, 32, 32, 32, 32, 32, 32]
}
df = pd.DataFrame(csv, columns= ['Temperature', 'Humidity', 'Latitude', 'Longitude'])
df.to_csv (r'C:\Users\Ron\Desktop\export_dataframe.csv', index = False, header=True)
print (df)The latitude and longitude data of a plant in an array will be the same and the temperature and humidity data will change over time. This was an example of the data plotted to csv with the help of pre-defined variables.
Finally, the plot looks as follows:
The purple bar shows the humidity percentage while the blue bar and white bar show the rate of temperature of an array. I have applied various filters for visualizing the trend in data even further like date-time wise data, trends in temperature data, trends in humidity data which can be viewed on the left bar
Demonstration Video:
Link to the Kepler Map plot:
Limitations:
To find the diseases in the Apple plant, Image processing and Classification is used. Sun light and angle of Image capture is the main factor which affects the classification parameter. For this, a case study of a farm is required. During a case study, I can capture Plant diseases from different angles and different saturation and contrast levels, along with different exposure and different background. Training the model with a complete dataset including all these parameters, will make the model accurate enough and easily deployable to classify unknown data. During night time, capturing classification of images based on a RGB Model cannot classify Images properly. For this purpose, I am training the model further with a GrayScale Dataset of images with captured data under NightVision Camera. Along with this, I am training the model further with images from different angles to predict and classify a disease from different planes. Along with the OpenVino model Optimization, I am further converting the RGB model to Gray colour space using the OpenVino Perprocessors Model Optimization - (Currently under progress). This is done using rgb_to_gray preprocessor
This shows image capture using infrared thermal camera's used to take in data during the night. The images in this are in the form of grayscale and hence, the model needs to be trained using this dataset to classify and process night time data.
Go to Market, Practicality and Viability:
- This model is deployed on the edge for processing and classification of Apple diseases. This Classification on the edge using OpenVino and NCS2 optimized models makes it available for classification of large video data on the edge in real time. Transferring Large amount of Video data to cloud for computation is not possible. This method is extremely expensive and also needs high end Internet Bandwidth in the farm. Neglecting these barriers, the Raspberry pi and NCS2 are able to perform plant disease classification on the edge.
- Real time Sensor data is available to the Farm owner. Hence the farm owner has Moisture data of the soil to check Moisture levels of the soil in real time. Climatic conditions like humidity and temperature are also available. Hence the farm owner has temperature and Humidity data of trees in Individual arrays.
- Streamlit web-front-end data which is available to get real time notifications on the Computer and even Smart phones. The farmer gets real time notifications of a particular plant in an array he wants to observe. He can see the real time Diseases detected in the Apple plant, real time moisture of the soil and humidity and temperature notifications of any specific plant. This data is available in the form of timely trends and the farmer can observe yearly trends in the data, monthly trends , daily trends or hourly trends of the data.
- On crossing a certain threshold or dropping below a threshold, the autonomous peristaltic pump is triggered to water the plants autonomously. The pump is also aware of the timings when the plant was last watered and the farmer gets a document to view all the times when the plant was watered. Besides this, a manual button to trigger the watering of the plants on the web app is also available.
- The farmer gets a Geo-Spatial Analysis of this data and he can view exactly the coordinates of the plants where the moisture level is low or where the humidity is high. This is performed through Satellite Imaging of the data and so the farm owner can view each plant data individually in the form of a Satellite view.
- The complete Apple farm management system is automated and Data of the Farm which was not available once, is made available using this method.
Hyperspectral deals with imaging narrow spectral bands over a continuous spectral range, producing the spectral fingerprint of all pixels in the scene. A sensor with only 20 bands can also be hyperspectral when it covers the range from 500 to 700 nm with 20 bands each 10 nm wide. (While a sensor with 20 discrete bands covering the VIS, NIR, SWIR, MWIR, and LWIR would be considered multispectral.)
These sensors can detect and identify minerals, vegetation and other materials that are not identifiable by other sensors. They are used in plant nutrient status, plant disease identification, water quality assessment, foliar chemistry, mineral and surface chemical composition and spectral index research.
Hyperspectral sensors have two methods for scanning: Push Broom and Whisk Broom.
Push Broom scanning, which is ideal for drone use, captures full spectral data simultaneously as the drone moves forward using a line of sensors that runs perpendicular to the drone's flight direction. With this method, the scanner is able to look at a particular area for a longer period of time enabling more light to be gathered.
Drone functioning in precision agriculture:
The basic architecture of a drone, without considering the payload sensors, consists of: (i) frame, (ii) brush-less motors, (iii) Electronic Speed Control (ESC) modules, (iv) a control board, (v) an Inertial Navigation System (INS), and (vi) transmitter and receiver. In precision agriculture, the drones are semi-autonomous. In that case, the drone has to fly according to the definition of a flight path in terms of waypoints and flight altitude. Thus, the drone has to embed on board a positioning measurement system (e.g., Global Navigation Satellite System, GNSS) for knowing its position with respect to the waypoints.
The payload of a drone includes all the sensors and actuators that are not used for the control of its flight (e.g., the gimbal with the RGB camera). In case of precision agriculture, the sensors embedded on drones are: multispectral camera, thermal camera, RGB camera
During collecting of data from a high altitiude, using hyperspectral and multispectral imaging, there a lot of atomsphere factors which affect the data. For this, the model needs atmospheric compensation and reduction of noise from the perceived data to predcit and analyse diseases in apple plants, This will be a future improvement after the first succesful deployement of the model.
This is the data view from an altitude of 50ft which leads to additional aggregation of atmospheric noise. To reduce this, the drone will be deployed at an altitude of 10ft to get a better accuracy while capturing the diseases.
Usually, for agriculture the terrain is scanned by using satellites with multispectral and thermal cameras. For precision agriculture, due to the needed high spatial resolution, drones are more suitable platforms than satellites for scanning. They offer much greater flexibility in mission planning than satellites. The drone multispectral and thermal sensors simultaneously sample spectral wavebands over a large area in a ground-based scene. After post-processing, each pixel in the resulting image contains a sampled spectral measurement of the reflectance, which can be interpreted to identify the material present in the scene. In precision agriculture, from the reflectance measurements, it is possible to quantify the chlorophyll absorption, pesticides absorption, water deficiency, nutrient stress or diseases. As a prototype, I have aimed to develop this model to predict diseases inclusively adding chlorophyll absorption prediction model in the second version of the prototype.
Adding GPS calibration on Drone for drone navigation:
Most GPS modules communicate with the Raspberry Pi via a simple serial connection. They send strings that contain GPS data and other status messages. These strings are called NMEA sentences.
Continuous location updating:
Adding location plotting on geospatial map for tracking the drone:
Tracking the Drone Altitude to maintain drone stability:
Functioning of the Technology:
Each time the RGB imaging camera captures the data and sends to the Raspberry Pi and NCS2, it classifies the data and sends the output. In order to know the location where the data was detected, for a plant in an array and accurately know the exact location of the diseased plant, GPS Calibration is important. For this purpose, a simpe python logic system needs to be deployed to the existing Jupyter Notebook to store the latitude and longitude of the classified data.
Each time a disease is detected, it stores the location of the data through a script as follows:
import serial #import serial pacakge from time import sleep import webbrowser #import package for opening link in browser import sys #import system package def GPS_Info(): global NMEA_buff global lat_in_degrees global long_in_degrees nmea_time = [] nmea_latitude = [] nmea_longitude = [] nmea_time = NMEA_buff[0] #extract time from GPGGA string nmea_latitude = NMEA_buff[1] #extract latitude from GPGGA string nmea_longitude = NMEA_buff[3] #extract longitude from GPGGA string print("NMEA Time: ", nmea_time,'\n') print ("NMEA Latitude:", nmea_latitude,"NMEA Longitude:", nmea_longitude,'\n') lat = float(nmea_latitude) #convert string into float for calculation longi = float(nmea_longitude) #convertr string into float for calculation lat_in_degrees = convert_to_degrees(lat) #get latitude in degree decimal format long_in_degrees = convert_to_degrees(longi) #get longitude in degree decimal format #convert raw NMEA string into degree decimal format def convert_to_degrees(raw_value): decimal_value = raw_value/100.00 degrees = int(decimal_value) mm_mmmm = (decimal_value - int(decimal_value))/0.6 position = degrees + mm_mmmm position = "%.4f" %(position) return position gpgga_info = "$GPGGA,"ser = serial.Serial ("/dev/ttyS0") #Open port with baud rateGPGGA_buffer = 0NMEA_buff = 0lat_in_degrees = 0long_in_degrees = 0 try: while True: received_data = (str)(ser.readline()) #read NMEA string received GPGGA_data_available = received_data.find(gpgga_info) #check for NMEA GPGGA string if (GPGGA_data_available>0): GPGGA_buffer = received_data.split("$GPGGA,",1)[1] #store data coming after "$GPGGA," string NMEA_buff = (GPGGA_buffer.split(',')) #store comma separated data in buffer GPS_Info() #get time, latitude, longitude print("lat in degrees:", lat_in_degrees," long in degree: ", long_in_degrees, '\n') map_link = 'http://maps.google.com/?q=' + lat_in_degrees + ',' + long_in_degrees #create link to plot location on Google map print("<<<<<<<<press ctrl+c to plot location on google maps>>>>>>\n") #press ctrl+c to plot on map and exit print("------------------------------------------------------------\n") except KeyboardInterrupt: webbrowser.open(map_link) #open current position information in google map sys.exit(0)This code gives the output as follows and prints the latitude and longitude of the region the GPS Module moves through. This gives the output as:
print("lat in degrees:", lat_in_degrees," long in degree: ", long_in_degrees, '\n')The above script prints the latitude and longitude of the region which can be further used to store in a csv file and the map can be deployed on kepler.gl map viewer to visualise the plot through geospatial analysis.
allowed_class == 'alternaria'
for allowed_class in class_names:
alternaria = 0
if alternaria = 0:
print("lat in degrees:", lat_in_degrees," long in degree: ", long_in_degrees, '\n')Additionaly, a functionality to store these outputs in the form of CSV can be added to make a fully funtional drone based mapping.
Why is drone based disease detection system more useful than individually deploying image sensors in each array?
Deploying Individual sensors in each array increases the cost by a significant amount. To reduce thi cost and make this product commercially viable, it is necessary to make an affordable working logic system. For this pupose, Drone based multispectral imaging with GPS Calibration proves to be a viable solution. In this system, the mapping of the complete farm plot can be performed using Drones along with Disease detection of the plants in the farm. The detecteion can also be done using a top down view system and also can simulate the mapping of the drone using a pre defined path. While deploying the model to inference on the drone, a high FPS based classification is important. Here the OpenVino optimization along with NCS2 is the key system that helps in classification on Raspverry Pi.
Leaf, Apple and Crop Area Index Estimation:LAI is a measure for the total area of leaves per unit ground area and directly related to the amount of light that can be intercepted by plants. It is an important variable used to predict photosynthetic primary production, evapotranspiration and as a reference tool for crop growth.
Usually this method is used for leaf area estimation but to simplify the calculations and the weight of model deployed in order to speed up the inferencing, an alternate algorithm has been developed by me which I am implementing upon and hence I've mentioned this in the future improvements,
The above image shows that a large number of apples are detected in the image but estimating the total yield of these is not possible by counting the number of estimated apples. For this purpose, calculating the total area of the detected apples is important.
This shows one of the apples detected in the classifications. For each detection, the xmin, xmax, ymin and ymax coordinates in the detections are stored. After taking these coordinates, estimating the box area is easy. In this way by estimating the box area of the apples the yield of the apples can be detected. Similarly, if diseases apples are detected, the yield is counted as a negative integer based on the magnitude of the estimation.
Logic used in yield estimation method:
Once the apple have been detected, it basically has 4 numbers describing the box: its top-left corner (x1,y1) and its bottom-right corner (x2,y2). Given those, we can also easily compute the area of the rectangle. To compute the area, we only need to compute the width as (x2-x1) and the height as (y2-y1) and multiply them.
The crop yield is also dependent on the variable: Distance between the camera and the crop. Based on this variables, the threshold to estimate the area and declare it's yield is set and accordingly the output of the yield volume is declared.
def compute_yield((x1,y1), (x2,y2)):
area, center = compute_area_and_center((x1,y1), (x2, y2))
volume = 0
if area > 100 and area < 150:
volume == 1 #in kilograms
if area > 151 and area < 250:
volume == 1.5Using this logic, the area of the crop and apple plant can be easily counted and the volume of the individual apple can be declared so that the far owner can have an estimation of the yield in his farm and similarly can estimate his yield for the quarter year.
Crop Yield, Climate and Rainfall prediction using OpenVino:Future Improvement:
Crop yield is a highly complex trait determined by multiple factors such as genotype, environment, and their interactions. Accurate yield prediction requires fundamental understanding of the functional relationship between yield and these interactive factors, and to reveal such relationship requires both comprehensive datasets and powerful algorithms.
Many studies have focused on explaining the phenotype (such as yield) as an explicit function of the genotype (G), environment (E), and their interactions (G × E). One of the straightforward and common methods was to consider only additive effects of G and E and treat their interactions as noise. A popular approach to study the G × E effect was to identify the effects and interactions of mega environments rather than more detailed environmental components. Several studies proposed to cluster the environments based on discovered drivers of G × E interactions used the sites regression and the shifted multiplicative models for G × E interaction analysis by dividing environments into similar groups. Burgueño et al. (2008) proposed an integrated approach of factor analytic (FA) and linear mixed models to cluster environments and genotypes and detect their interactions. They also stated that FA model can improve predictability up to 6% when there were complex G × E patterns in the data . Linear mixed models have also been used to study both additive and interactive effects of individual genes and environments
More recently, machine learning techniques have been applied for crop yield prediction, including multivariate regression, decision tree, association rule mining, and artificial neural networks. A salient feature of machine learning models is that they treat the output (crop yield) as an implicit function of the input variables (genes and environmental components), which could be a highly non-linear and complex function. Liu et al employed a neural network with one hidden layer to predict corn yield using input data on soil, weather, and management. Drummond et al. used stepwise multiple linear regression, projection pursuit regression, and neural networks to predict crop yield, and they found that their neural network model outperformed the other two methods. Marko et al. proposed weighted histograms regression to predict the yield of different soybean varieties, which demonstrated superior performances over conventional regression algorithms.
Weather Prediction
Weather prediction is an inevitable part of crop yield prediction, because weather plays an important role in yield prediction but it is unknown a priori.
The reason for using neural networks for weather prediction is that neural networks can capture the nonlinearities, which exist in the nature of weather data, and they learn these nonlinearities from data without requiring the nonlinear model to be specified before estimation (Abhishek et al., 2012). Similar neural network approaches have also been used for other weather prediction studies.
Yield Prediction Using Deep Neural Networks:
We can train two deep neural networks, one for yield and the other for check yield, and then we can use the difference of their outputs as the prediction for yield difference. This model structure was found to be more effective than using one single neural network for yield difference, because the genotype and environment effects are more directly related to the yield and check yield than their difference.
Deep neural network structure for yield or check yield prediction. The input layer takes in genotype data (G∈ ℤn×p), weather data (W∈Rn×k1W∈ℝn×k1), and soil data (S∈Rn×k2S∈ℝn×k2) as input. Here, n is the number of observations, p is the number of genetic markers, k1 is the number of weather components, and k2 is the number of soil conditions. Odd numbered layers have a residual shortcut connection which skips one layer. Each sample is fed to the network as a vector with dimension of Rp+k1+k2ℝp+k1+k2.
To predict the output for weather, rainfall and crop yield, designing the neural network that has appropraite number of layers and generates the expected output is important. For this viewing and exploring the neural network is necessary. Here, OpenVino plays an important role to visualise these layers and also adding custom layers which is not possible using keras.
Custom Layer Operation
- Responsible for specifying the attributes that are supported by the custom layer and computing the output shape for each instance of the custom layer from its parameters.
The--mo-opcommand-line argument shown in the examples below generates a custom layer operation for the Model Optimizer. - Custom Layer Operation
Responsible for specifying the attributes that are supported by the custom layer and computing the output shape for each instance of the custom layer from its parameters.
The--mo-opcommand-line argument shown in the examples below generates a custom layer operation for the Model Optimizer.
The Decision tree classifiers uses greedy approach hence an attribute chooses at first step can’t be used anymore which can give better classification if used in later steps. Also it overfit the training data which can give poor results for unseen data. So, to overcome this limitation ensemble model is used. In ensemble model results from different models are combined. The result obtained from an ensemble model is usually better than the result from any one of individual models
The probability density functions of the ground truth yield and the predicted yield by DNN model. The plots indicate that DNN model can approximately preserve the distributional properties of the ground truth yield.
Predicted Yield Outputs:
Importance Comparison Between Genotype and Environment
To compare the individual importance of genotype, soil and weather components in the yield prediction, we obtained the yield prediction results using following models:
DNN(G): This model uses the DNN model to predict the phenotype based on the genotype data (without using the environment data), which is able to capture linear and nonlinear effects of genetic markers.
DNN(S): This model uses the DNN model to predict the phenotype based on the soil data (without using the genotype and weather data), which is able to capture linear and nonlinear effects of soil conditions.
DNN(W): This model uses the DNN model to predict the phenotype based on the weather data (without using the genotype and soil data), which is able to capture linear and nonlinear effects of weather components.
Genotype and environment data are often represented by many variables, which do not have equal effect or importance in yield prediction. As such, it is vital to find important variables and omit the other redundant ones which may decrease the accuracy of predictive models. In this paper, we used guided backpropagation method which backpropagates the positive gradients to find input variables which maximize the activation of our interested neurons.
Bar plot of estimated effects of 8 soil conditions. AWC, OM, CEC, and KSAT stand for available water capacity, organic matter, cation exchange capacity, and saturated hydraulic conductivity, respectively. The Bar plot indicates that percentage of clay and soil pH were more important than the other soil conditions.
Bar plot of estimated effects of 6 weather components measured for 12 months of each year, starting from January. The vertical axes were normalized across all weather components to make the effects comparable.
Thank you for viewing my project














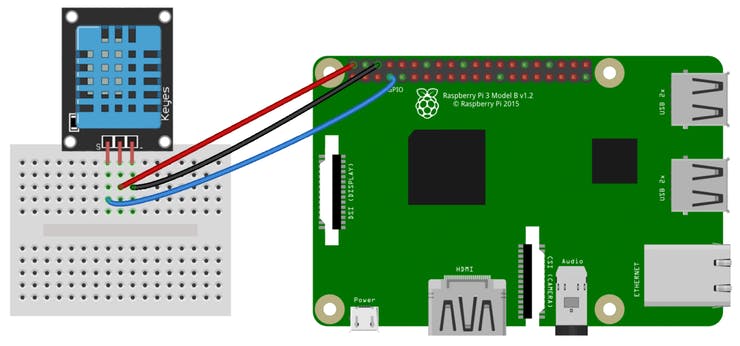
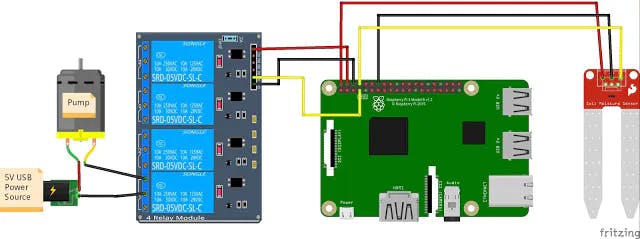









_3u05Tpwasz.png?auto=compress%2Cformat&w=40&h=40&fit=fillmax&bg=fff&dpr=2)

Comments
Please log in or sign up to comment.