Covid-19 pandemic has devastated social life and damaged the economy of the global population with a constantly increasing number of cases and fatalities each day. A critical step in the fight against COVID-19 is the effective screening of infected patients, with one of the key screening approaches being radiology examination using chest radiography (.i.e. X-Ray images).
Detecting COVID-19 infections on Chest X-Rays with high accuracy and speed may help quarantine high risk patients while test results are awaited with faster diagnosis!
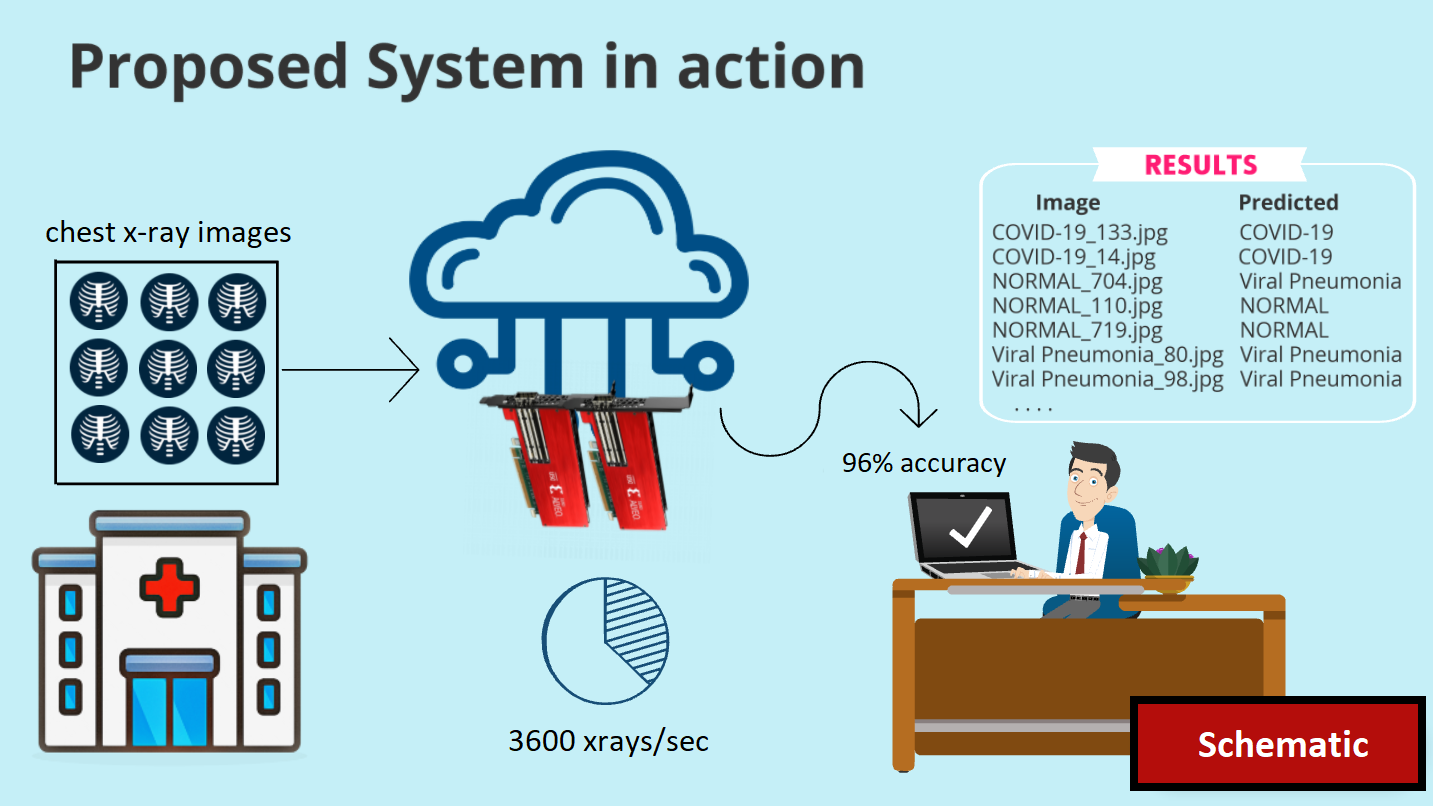
In this scheme, we propose a highly accurate application that can classify X-Ray images in 3 classes (COVID, NORMAL, Viral Pneumonia) using Neural Networks on FPGAs with great speed as well.
The CNN model was trained with hardware-aware optimizations then was quantized to 8-bit and finally compiled to run on Xilinx Alveo U50 FPGA through Vitis AI. To the best of our knowledge, this application has not yet been considered for cloud FPGAs while the accuracy and speed achieved surpasses any previous known implementation of CNNs for X-Ray Covid detection. Specifically, it can classify X-Ray images at a rate of 3600 FPS with 97% accuracy and a speed-up of 17.6x vs CPU and 3x vs GPU.
The models provided are not intended for self-diagnosis and anyone should seek help from their local health authorities if needed.
The theoryRadiography examination is a very efficient method by radiologists to look for visual indicators associated with SARS-CoV-2 viral infection (e.g., chest X-ray (CXR) or computed tomography (CT) imaging). X-Ray imaging has several advantages over other diagnostic tests as it is much more widespread, cost effective, and portable as opposed for example to CT-scanners.
In this scheme, Convolutional Neural Networks (CNNs) can automate the diagnosis using Deep Learning approaches and revolutionize medical processes especially nowadays.
However, CNN models have a large number of parameters as well as high hardware requirements and companies now might require a demanding workload of many terabytes of data or more to be processed every day. FPGAs can contribute fundamentally as is it shown that they have been extremely effective on CNN tasks due to their massive parallelism and reconfigurability on the bit level while having high power efficiency which is essential for cloud workloads.
Getting startedIn order to deploy and test the models' inference on FPGA you must prepare your machine by following the next steps. The project is also provided in this Github repo. You will need:
- An Ubuntu host PC (16.04 or 18.04)
- Setup Alveo U50 FPGA Card if you haven't already. Install XRT and Alveo target platform files. Follow instructions
- Install Docker and ensure your linux user is in the docker group:
sudo apt install docker -y
sudo groupadd docker
sudo usermod -aG docker $USER
newgrp docker- Download our github repo (~70MB in size)
git clone https://github.com/dimdano/Covid4HPC.git- Pull and run Vitis AI prebuilt docker (release 1.2.82 recommended).
chmod +x docker_run.sh
./docker_run.sh xilinx/vitis-ai:latest- Install Xilinx DPU IP for the Alveo U50. While inside docker run:
wget https://www.xilinx.com/bin/public/openDownload?filename=alveo_xclbin-1.2.1.tar.gz -O alveo_xclbin-1.2.1.tar.gz
tar xfz alveo_xclbin-1.2.1.tar.gz
sudo cp alveo_xclbin-1.2.1/U50/6E300M/* /usr/lib- You can head to
FPGA_demoto test the ready-to-run application. For the other tutorials you will need to download COVID-19 Radiography dataset from here. Then place the 3 class folders (Covid, Normal, Viral Pneumonia) inside thedatasetfolder of the repo.
Our aim was to produce a very efficient and accurate CNN architecture tailored for the detection of COVID-19 cases from X-Ray images that is also deployable for FPGA acceleration through Vitis AI. In this section we will briefly show you how we implemented the CNN models.
- Preprocess: First, we downloaded the dataset which was COVID-19 Radiography Database from Kaggle [link]. It consists of 219 COVID-19 positive images, 1341 normal images and 1345 viral pneuomonia images. We pre-processed the data by resizing the images to the typical 224x224 dimensions found in most CNNs, with 25% of the dataset for validation.
-----------------COVID------NORMAL----VIRAL PNEUMONIA--
========================================================
X-Ray Images: 219 1341 1345- Train: We designed 2 different models in Tensorflow/Keras framework with different topologies; a Custom CNN [topology] and a DenseNetX [topology] type model. The pretrained Keras models can be found [here].
The models were carefully designed in order to compile efficiently for DPUv3E accelerator IP targeting the Alveo U50. Thus, many hardware-oriented optimizations/considerations took place on the topology of the Neural Networks such as type of layers, size of kernels, etc. such as below:
tf.keras.layers.Conv2D(64, 3, strides = 1, padding='same'),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.Activation('relu'),Also, we used data augmentation of the training data to increase the diversity of the images as seen here (zooming, rotating, etc.).
train_aug = ImageDataGenerator(height_shift_range = 0.1,
width_shift_range=0.1,
rotation_range=10,
horizontal_flip=True,
shear_range = 0.1,
zoom_range=0.1)Last, another improvement that we did was to impose class weights for the imbalanced Covid class so as to have a more balanced training.
class_weight = {0: 6, 1: 1, 2: 1}For the deployment and acceleration of our trained CNNs on the Alveo U50 FPGA card we used Xilinx's Vitis AI. It is a development stack for AI inference on Xilinx hardware platforms that consists of optimized IP, tools and libraries.
You can follow the next steps to easily reproduce and run our HW-model for FPGA deployment or you can head to FPGA demo section to test the ready-to-run application instantly. Make sure you have completed the Setup instructions first and you are in Vitis AI docker.
The tutorial runs for "CustomCNN" model by default. If you would like to use the DenseNetX model run source ./0_setenv.sh DenseNetX instead of source ./0_setenv.sh.
1. First run the following script to set variables and activate vitis-ai-tensorflow
source ./0_setenv.sh2. Convert the model to TF Frozen Graph by running the following command:
source ./1_keras2tf.sh3. Evaluate the prediction accuracy on the 32-bit model.
source ./2_eval_frozen.sh4. Quantize from 32-bit floating point to 8-bit fixed point with calibration images:
source ./3_quant.sh5. Evaluate the prediction accuracy of the quantized 8-bit model:
source ./4_eval_quant.sh6. Compile the quantized model and generate the xmodel file for the target FPGA (Alveo U50):
source ./5_compile.sh7. Prepare the application folder and generate the test images:
source ./6_make_target.sh8. Go to build/target/ folder and run application with:
/usr/bin/python3 run_inference.py -m model_dir/fpga_model.xmodel -t 8You can select model with -m option or the number of threads with -t option.
Note* The DPU runs at 300MHz. If you experience issues with the FPGA becoming unstable, it might need reset. You can scale down the frequency to overcome these issues. For example run the following to reduce to 250Mhz:
/opt/xilinx/xrt/bin/xbutil program -p /usr/lib/dpu.xclbin
/opt/xilinx/xrt/bin/xbutil clock -d0 -g 83If you want to test the ready-to-run application head to FPGA_demo/ folder and run the following command to test the "CustomCNN" model on FPGA:
/usr/bin/python3 run_inference.py -m model_dir/CustomCNN.xmodel -t 8The FPGA will run the model to predict the X-Ray images stored in the images/ folder. The Frames/Sec (FPS) and Accuracy achieved will be shown. You can select the model to infer with -m option or the number of threads with -t option. (If you experience issues with FPGA see Note* )
The results are saved in a ".txt" file which includes the predicted results of the all the input images as seen (partially) below.
The results can be easily transfered remotely to a hospital or clinic so that doctors can assess better the condition of the patients.
ResultsA jupyter notebook cpu-evaluation.ipynb is provided in the repo to evaluate the model on the following classification metrics. Also, you can visualize the regions of attention of the X-ray images.
First source Vitis AI environment:
source ./0_setenv.shFor the visualization package only, you will need to install a higher TF-version and tf_keras_vis package by running:
pip install tensorflow==2.0.2 tf_keras_visThen, while inside the docker container you can run jupyter (port 8888 enabled in host):
jupyter notebook --no-browser --port=8888 --allow-rootNext, copy the URL shown afterwards in a host browser to run jupyter.
- Classification Report : For the evaluation of the model we ran a classification report. The best model, which was the "CustomCNN", achieved an overall accuracy of 97%.
Report precision recall f1-score Samples
===============================================================
COVID-19 1.00 0.95 0.97 55
NORMAL 0.98 0.96 0.97 335
Viral Pneumonia 0.96 0.98 0.97 337
accuracy 0.97 727
macro avg 0.98 0.96 0.97 727
weighted avg 0.97 0.97 0.97 727- Confusion Matrix: The predicted images can be also visualized in a confusion matrix. Each row shows the instances in an actual class while each column represents the instances in a predicted class.
- Visualizations: Another way to demonstrate the results in the qualitative comparison, was to generate saliency maps for our model’s predictions using tf-keras-vis package [link]. These visualizations are very important because they give us an insight on the model's classifier capability as well as validate the regions of attention.
- Inference Speed: We measured the performance when running the FPGA demo. We compared it with CPU and GPU architectures using the default settings in Tensorflow for model inference (float). Below are the results:
Architecture System Frames/Sec
===============================================================
CPU Intel Xeon Silver 4210 204
GPU Nvidia V100 1157
FPGA Xilinx Alveo U50 3606- Multi-class classification: Our model classifies 224x224 X-Ray images in 3 classes (COVID, NORMAL, Viral Pneumonia) as opposed to other works which only do binary classification on COVID.
- High accuracy: The CustomCNN model provided achieves 96% accuracy (quantized) on FPGA. It overperforms other CNN models from previous work on Covid-19 detection. (see this table for comparison which is from a paper published in "Computers in Biology and Medicine" journal)
- Small and efficient model: The trained model is very small (24MB) while the quantized model is (4.4MB). Also, the GFLOPs required for model inference is low (1.03G). This means that it does not have high memory or compute needs.
- High inference speed: The model performs very well in terms of throughput with a measured speed of 3600 FPS. It has a 17.6x speed-up from CPU (
Intel Xeon Silver 4210) and 3x speed-up from GPU (Nvidia V100) on the model inference on Keras. - Contribution to Xilinx Model Zoo: The CustomCNN model achieves the highest throughput and great GOPs efficiency between Alveo U50 models provided in Xilinx Model Zoo [link]. In addition, we provided a 2nd model based on DenseNetX topology which is also a nice contribution to the model zoo.
- Open source code: All the trained models, the steps to reproduce the FPGA application as well as the results are provided in our git repository.




Comments
Please log in or sign up to comment.