From medicine to beer, product of yeast cultures are an indispensable part of our daily lives, hence predictable control of gene expression is necessary for the rational design and optimization of yeast culture growth. In the yeast Saccharomyces cerevisiae, the promoter is one of the most important tools available for controlling gene expression. However, the complex expression patterns of yeast promoters have not been fully characterized and it is often difficult and time consuming to adjust the concentrations of activator compounds to achieve desirable levels of production (B. Peng et.al, 2015). Our team therefore suggests using alight-dependent promoter that takes concentration of desired product as the input argument and adjusts the red light to far red-light ratio appropriately to achieve optimum production rate.
Why have alight-adjustable promoter?1. Quick, easy, non-invasive way to switch gene expression on and off, which doesn’t require any chemicals.
2. Allows the concentration of an enzymes in the metabolic pathway to be optimized without re-engineering the circuit with a different promotor
3. Gives the flexibility to increase and decrease production in response to demand and is adaptable with organisms’ growth cycle.
4. Allows checkpoints to be used in synthesis e.g. ensures that a certain concentration of substrate is achieved before producing a desired concentration of enzyme – essentially using the system to maximize the efficiency of a synthesis which reduces, as far as possible, the fitness cost of the system on the organism.
5. Can be used to differentiate parts of a plate i.e. creating spatial sub populations (create sub populations expressing different genes or different amount of a gene in different areas).
6. Eventually if multiple different colored-light adjustable promoters can be created then the concentration of each of the compounds in the steps of a synthesis can be controlled and optimized using light, without re engineering the promoters.
Project outlineThe basis of our project is a light adjustable promotor created by TU Munich’s 2012 iGEM team named Brew. This promoter utilizes a Reverse Yeast-Two Hybrid Based Light-Switchable Promoter System which is based on the yeast two-hybrid screen to identify protein-proteininteraction. Instead of the normal conjugated proteins in the screen, it uses the photo-convertible binding of PhyB to PIF3 to create a light adjustable promoter. Although this system was originally used as an on-off switch for transcription, it has the capability to up regulate and down regulate expression. This can be done through altering the light intensity, ratio of red and far-red light, or by flashes of red and far-red light.
Our project attempts to model this adjustable promoter in various synthetic systems in yeast and create a node in XOD that works as a kind of potentiometer for the synthetic circuit. The XOD node can receive different sensor inputs like chemical sensor inputs and outputs an LED response that can alter gene expression in the desired way to achieve a set target.
An XOD node was developed with sensors as inputs, a target for expression, and LED’s as outputs. The node is capable of receiving chemical concentration input from a culture of Saccharomyces cerevisiae at regular intervals and comparing it against a stored set value. If a mismatch is present between the measured and set values, the algorithm can optimize the LED output until the desired target is achieved.
• Main Patch
Figure 2 shows the main patch for the XOD node. The function of this patch is to allow the user to maintain optimal concentration of the desired product in the culture in one of the three ways:
1. Changing Light intensity
2. Changing duration of light exposure
3. Changing wavelength of light
The Boolean input true at the top left-hand corner acts as a switch. This allows the user to turn the system on or off.
• Patch 2
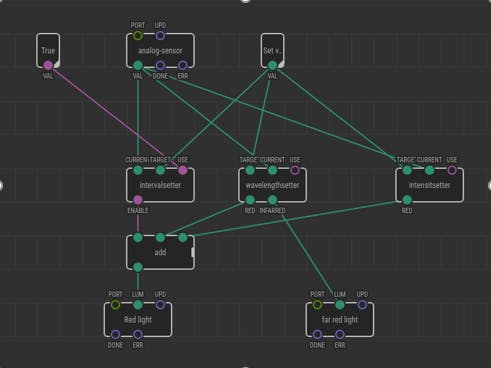
This patch accepts two input arguments: current value of the concentration of the compound and the target value of the concentration of the compound. There are two delay functions present. The delay on the left is to set the time between the intervals, while the delay on the right is to set the time duration for which the light is turned on. When the system finds that the concentration is less than the target value and the delay between the intervals has reached zero, then the light is turned on provided the condition that the entire system is turned on by user.
• Patch 3
The function of this patch is to turn on the far-red light when the concentration of the desired product exceeds the target value. Far-red light has an inhibitory effect on the system. On the other hand, red-light is turned on when the current value is less than the target value. In order to convert the input from the sensor into a Boolean form, the left hand side of the figure shows a possible mathematical manipulation which employs the use of arctan function because of its suitable properties for our system. The outputs as seen in the figure above are connected in such a way, that for most of the time, both the red and far-red light are on simultaneously. It is important to note that this mathematical manipulation of the input value is arbitrary. More rigorous calculations could be applied to process actual data obtained from a yeast culture.
• Patch 4
The main function of the patch 4 is to modulate the intensity of the red light. When the product concentration falls below the desired value, the red light is turned on by the patch 3. The patch 4 can then increase the intensity of the red light in order to achieve a higher production rate of the product.
The patch 4 utilizes similar mathematical methods methods as mentioned previously for the patch 3. There are two nested if-else functions within the patch 4, to ensure that the intensity of the red light increases only if the entire system is in on state and the concentration of the desired product is less than the target value.
Future work• Now that we have developed the XOD node, the first step on the biological side will be to replicate the promoter system using the same bio-bricks and components utilized by TUM iGEM team. This could then be tested in a simple synthesis system with red and far-red LEDS to show that it works to upregulate and down regulate gene expression. A simple chemical synthesis e.g. a two or three step process with our regulatable promoter could be implemented in the organism to test the input sensors used by the node.
• It will be interesting to investigate how the node works in a different system i.e. a different synthetic pathway. This can allow us to find whether the algorithm for LED outputs can be same or should change for the different system / different types of system.
• A hypothetical system in XOD with multiple of these nodes can be investigated. This possibility can also be explored by using multiple adjustable promoters e.g. using different colors.
References1. B.Peng, et.al, 2015, “Controlling heterologous gene expression in yeast cell factories on different carbon substrates and across the diauxic shift: a comparison of yeast promoter activities” PMCID: PMC4480987, [URL: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4480987/]
2. “Light Switchable Promoter”, 2012, iGEM team TMU. [URL: http://2012.igem.org/Team:TU_Munich/Project/Light_Switchable_Promoter]




Comments
Please log in or sign up to comment.