Phenotyping involves the quantitative assessment of the anatomical, biochemical, and physiological plant traits. Natural plant growth cycles can be extremely slow, hindering the experimental processes of phenotyping. Deep learning offers a great deal of support for automating and addressing key plant phenotyping research issues. Machine learning-based high-throughput phenotyping is a potential solution to the phenotyping bottleneck, promising to accelerate the experimental cycles within phenomic research.
The influence of climate change, and due to it's unpredictable nature, majority of agricultural crops have been affected in terms of production and maintenance. Hybrid and cos-effective crops are making their way into the market, but monitoring factors which affect the increase in yield of these crops, and conditions favorable for growth have to be manually monitored and structured to yield high throughput. Farmers are showing transition from traditional means to hydroponic systems for growing annual and perennial crops. These crop arrays possess growth patterns which depend on environmental growth conditions in the hydroponic units. Semi-autonomous systems which monitor these growth may prove to be beneficial, reduce costs and maintenance efforts, and also predict future yield beforehand to get an idea on how the crop would perform. These systems are also effective in understanding crop drools and wilt/diseases from visual systems and traits of plants.
Forecasting or predicting the crop yield well ahead of its harvest time would assist the strategists and farmers for taking suitable measures for selling and storage. Accurate prediction of crop development stages plays an important role in crop production management. Such predictions will also support the allied industries for strategizing the logistics of their business. Several means and approaches of predicting and demonstrating crop yields have been developed earlier with changing rate of success, as these don’t take into considerations the weather and its characteristics and are mostly empirical.
Crop yield estimation is also affected by taking into account a few other factors. Plant diseases enormously affect the agricultural crop production and quality with huge economic losses to the farmers and the country. This in turn increases the market price of crops and food, which increase the purchase burden of customers. Therefore, early identification and diagnosis of plant diseases at every stage of plant life cycle is a very critical approach to protect and increase the crop yield.
In this article, I propose an Embedded Machine Learning approach to predicting crop yield and biomass estimation of crops using an Image based Regression approach using EdgeImpulse that runs on Edge system, Sony Spresense, in real time. This utilizes few of the 6 Cortex M4F cores provided in the Sony Spresense board for Image processing, inferencing and predicting a regression output in real time. This system uses Image processing to analyze the plant in a semi-autonomous environment and predict the numerical serial of the biomass allocated to the plant growth. This numerical serial contains a threshold of biomass which is then predicted for the plant. The biomass output is then also processed through a linear regression model to analyze efficacy and compared with the ground truth to identify pattern of growth. The image Regression and linear regression model contribute to an algorithm which is finally used to test and predict biomass for each plant semi-autonomously.
IntroductionAdvancements in computer vision and machine learning technologies have transformed plant scientists ability to incorporate high-throughput phenotyping into plant breeding. Detailed phenotypic profiles of individual plants can be used to understand plant growth under different growing conditions. As a result, breeders can make rapid progress in plant trait analysis and selection under controlled and semi-controlled conditions, thus accelerating crop improvements. In contrast to existing invasive methods for accurate biomass calculation that rely on plant deconstruction, this system used non-invasive alternative are in commercial applications which leaves the crops intact. Unfortunately, current commercially available platforms are large and very expensive. The upfront investment limits breeders' use of high-throughput phenotyping in modern breeding programs.
For agricultural applications the biomass is a powerful index due to its immediate connection with the crops health condition and growth state. Predicting sequential biomass of plants may serve as important indices to co-relate environmental growth with crop biomass. This approach presents using economical and cost-effective methods to approximate biomass using a Regression approach in Computer Vision DNN models. The regression model uses 2 Dimensional Convolutional Layers. Vision based Regression models help not only in calculating mean difference and increase in biomass but also understand visual cues in plants and predict the biomass evolution based on such cues. The objective of such an approach is to enable temporal analysis of biomass from frames to allow adapting to the planned environment and factors objectively. To take into consideration, wilting leaves observed progressively over frames suggests a decrease in biomass of plant over time, and can be monitored semi autonomously in farms. While existing approaches involve unimplementable algorithms or intensive computation, costly hardware or offline or batch processing, which is delayed calculation, this approach attempts to be implementable and not stress on inefficacious data from plants or inference.
Taking One step further to fulfill UN Sustainable Development Goals -
This project aims to expand scope of UN's second SDG and entail few of the Goal Targets by bringing in semi-autonomous monitoring systems for food production monitoring and yield production methodology to "increase productivity and production by implementing resilient agricultural practice." as highlighted in second UN SDG goal target.
Material and MethodsData Accumulation :
Most of the dataset used to train this model was adopted from the Paper "Growth monitoring of greenhouse lettuce" by Lingxian Zhang et al. There were 3 kinds of datasets offered in this paper, one of them being the raw dataset curated under unmonitored sunlight conditions. The other dataset was an augmented version of the raw dataset synthesized and generated with all images having similar light, illuminance and saturation. The third dataset contains spatial and depth information of these plants under the same environment and observed growth patterns. In this approach, we'll be using the augmented dataset to increase efficacy of model and couple images in a similar visual pattern.
Greenhouse lettuce image collection and preprocessing -
The experiment was conducted at the experimental greenhouse of the Institute of Environment and Sustainable Development in Agriculture, Chinese Academy of Agricultural Sciences, Beijing, China (N39°57′, E116°19′). Three cultivars of greenhouse lettuce, i.e., Flandria, Tiberius, and Locarno, were grown under controlled climate conditions with 29/24 °C day/night temperatures and an average relative humidity of 58%. During the experiment, natural light was used for illumination, and a nutrient solution was circulated twice a day. The experiment was performed from April 22, 2019, to June 1, 2019. Six shelves were adopted in the experiment. Each shelf had a size of 3.48 × 0.6 m, and each lettuce cultivar occupied two shelves. [Ref]
The number of plants for each lettuce cultivar was 96, which were sequentially labeled. Image collection was performed using a low-cost Kinect 2.0 depth sensor. During the image collection, the sensor was mounted on a tripod at a distance of 78 cm to the ground and was oriented vertically downwards over the lettuce canopy to capture digital images and depth images. The original pixel resolutions of the digital images and depth images were 1920 × 1080 and 512 × 424, respectively. The digital images were stored in JPG format, while the depth images were stored in PNG format. The image collection was performed seven times 1 week after transplanting between 9:00 a.m. and 12:00 a.m. Finally, two image datasets were constructed, i.e., a digital image dataset containing 286 digital images and a depth image dataset containing 286 depth images. The number of digital images for Flandria, Tiberius, and Locarno was 96, 94 (two plants did not survive), and 96, respectively, and the number of depth images for the three cultivars was the same.
Since the original digital images of greenhouse lettuce contained an excess of background pixels, this study manually cropped images to eliminate the extra background pixels, after which images were uniformly adjusted to 900 × 900 pixel resolution.The Figure below shows examples of the cropped digital images for the three cultivars.
Prior to the construction of the CNN model, the original digital image dataset was divided into two datasets in a ratio of 8:2, i.e., a training dataset and a test dataset. The two datasets both covered all three cultivars and sampling intervals. The number of images for the training dataset was 229, where 20% of the images were randomly selected for the validation dataset. The test dataset contained 57 digital images. To enhance data diversity and prevent overfitting, a data augmentation method was used to enlarge the training dataset.. The augmentations were as follows: first, the images were rotated by 90°, 180°, and 270°, and then flipped horizontally and vertically. To adapt the CNN model to the changing illumination of the greenhouse, the images in the training dataset were converted to the HSV color space, and the brightness of the images was adjusted by changing the V channel. The brightness of the images was adjusted to 0.8, 0.9, 1.1, and 1.2 times that of the original images to simulate the change in daylight. [Ref]
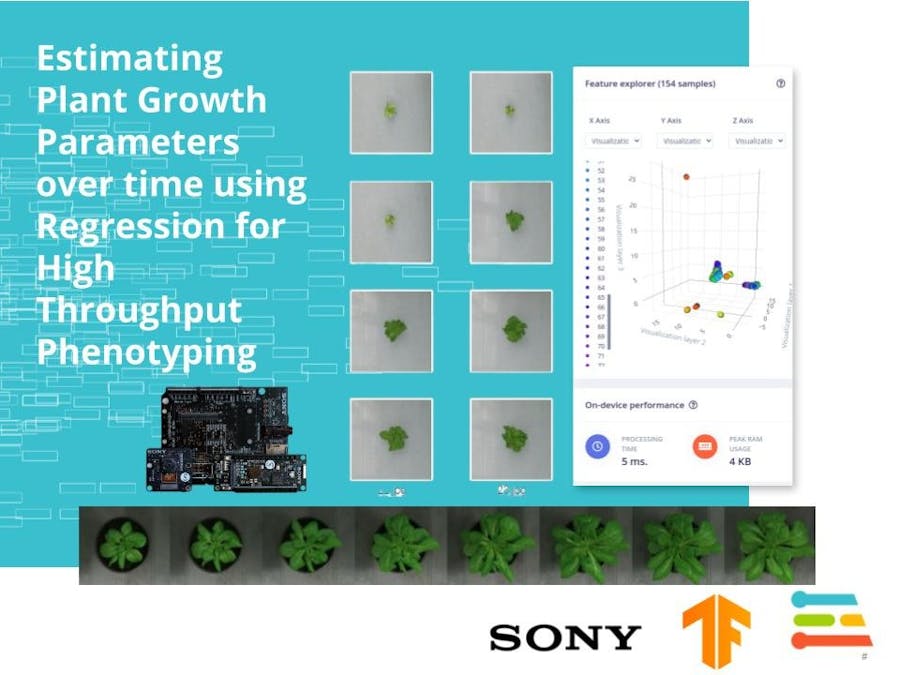
The raw dataset acquired was thus augmented and optimized to be fed into the Convolutional Neural Network for Regressional analysis. These aligned image pairs serve as input dataset in EdgeImpulse Studio. The below Figure illustrates how raw images perform as compared to Augmented ones with equalized lighting and saturation throughout the images. (Lettuce Flandria)
BelowFigure Illustrates Lettuce Tiberius Dataset-
The above figure demonstrates Synthesized Images for Neural Network possessing equalized illuminance, RGB Channels and saturation to maintain consistent Data input for CNNs.
The synthesized images were further augmented and a translational blur parameter of 0.01 was added to the images to be able to predict the biomass of lettuce even while the source from which the image is being captured is in motion. An example of translational motion blur are noticed on motorized plates or Machine Motion Drivers illustrated below -
The figure shown below demonstrates how the data for the Lettuce variants are captured and ingested. The camera angles were placed vertically perpendicular with respect to the ground plane and the distance between ground plane was adjusted to 78cm or 0.78m for each plant to ensure standardized images and ensure deviation in biomass increase and Leaf growth is in constant incrementation. The approximate image area covered by each cultivar is 4.176 meter squared and the approximate image area covered by each lettuce shoot is 0.0435 meter squared. The images captured are standardized to 128*128 pixels to make it easier for DNNs to scale and process. This implies that each 128*128 pixel image occupies 0.0435 meter squared or 435 cm squared area. Each pixel would hence occupy 2.65 x 10^-2 cm squared area. This conversion to ground scale is essential for computing not only relative but also absolute Leaf Area Index and Biomass for each plant predicted and verify it with ground truth. The process below has to be imitated while capturing test images or if the laboratory conditions vary, then adjust Field of Object in the image frame and either zoom in or zoom out corresponding to distance between/ depth of camera and plant.
Embedded Inference Board used for Real time processing:
The Sony Spresense Neural Inference board, main board and Camera system:
The Sony Spresense is a suite of Embedded systems mainly suited and widely used for Vision based solutions consisting image classification, inferencing and regression based prediction. It is ideally suited for low power, real time inferencing applications, suited for this system. The Sony Spresense comes with on-board 1536 kB RAM and 8192 kB ROM for Inference. The system allows processing multiple pipelines and DNNs/CNNs withing the data range. The main board itself is compact enough to be dropped into many production-grade systems without much fuss, and from a software standpoint, there are several options available, from using C/C++ SDK or Python API, whichever goes best with the system. This application features using pre-compiled C++ binaries for the system from EdgeImpulse Studio, leaving compilation headaches to the EdgeImpulse compiling studio.
The proceeding diagram demonstrates test data accumulation and live classification setup using Sony Spresense which will be elaborated in the latter part.
Ingesting the Cultivar Dataset to EdgeImpulse Studio for pre-processing and Feature Extraction:
For starters, EdgeImpulse has provided more details on how to get started with EdgeImpulse in their docs section - https://docs.edgeimpulse.com/docs/getting-started
There are two methods of data upload on the EdgeImpulse Studio, one of them including the EdgeImpulse Ingestion uploader API - https://docs.edgeimpulse.com/docs/cli-uploader
and the other being the visual uploader which in most cases is more preferred, for ease and simplicity. The figure demonstrates visual data ingestion process.
Here, the infer from filename selection is used for creating a regression dataset. Regression dataset classes are numerically named in progression of images to be fed in the CNN. The CNN uses numerical class interpolation instead of class name to train the regression model.
The dashboard sorts and orders data and is displayed with number of classes, image signature and label and a few filters to sort dataset.
Thereafter, an impulse is created in the impulse design tab with the necessary input block, processing block, learning block and output features.
The parameters block takes in pixel-by-pixel input features and converts them to scalar Raw features which can be interpreted by the CNN.
For each image processed, it consists of a feature and EdgeImpulse Studio projects a plot of these features in terms of 3 Visualization layers. A local visualization of the learned features is plotted which explains the pixels important for classification.
The above image illustrates the Architecture of the CNN Model. It consists of two 2D Convolutional Layers, two pooling layers and one Fully Connected Layer (FCN).
The CNN took pre-processed, 3 Channel images of Greenhouse Lettuce of size 128x128 after the feature-extraction was completed. Each convolution layers was capped with Kernels of size 3x3 which were used to extract the features. The Max Pooling Layers in the CNN were adapted with Kernels of size 2x2 and stride of 2, which is set to default in most CNNs. The CNNs were equipped with Max Pooling Function instead of an average polling function. A Dropout of rate 0.25 was used in the CNN to stabilize the rate of Dropout and not exceed to a higher rate of 0.5. A constant Learning Rate of 0.002 was set in the model, and the Batch Size was reduced to 16, which performed marginally better in comparison to 32. The Neural Network was trained and capped at 225 epochs, after which the loss function started peaking again. Mean Squared Error function was used to evaluate the loss function in the model. A few other hyperparameters were adjusted and evaluated to increase the model efficacy.
EdgeImpulse Studio provides default CNN Architectures selected for the Regression Model which are smaller and allow real time performance. If you wish to edit few of the Architectural layers, you can do so in the EdgeImpulse Studio.
Two models were subsequently trained to compare performance and loss rates between change in parameters used for labels. For the first model, the actual Leaf Area Index(LAI) (Ground Truth) Calculated via segmentation of the plant was used as labels. These are rational, decimal numbers like 77.8135, 90.4532 in cm^2 units.
The second model included Day-wise labels corresponding to growth stages of the plant. Eg- for data captured on Day 1 - label 1 was used (unitless integral vales). The second model did not show complete consistency in plant growth progression and hence the model performed poor and had larger weights and features to learn from.
To my surprise, the model which contained labels in terms of LAI achieved near stellar accuracy of 0.51(MSE - Mean Squared Error), and the second model was much heavier, slow in inference and also had a high loss function of 14.71.
The Regression model performed significantly better as compared to usual Regression models which peak at a loss rate of ~ 100. The loss function is calculated using Mean Squared Error gradient. The Epochs were set to 130 for training, while the learning rate to 0.005, which allowed faster learning and better results. The model loss stabilized after 35 epochs after which it continuously converged to a plateau descent, and remained stable for the rest. Comparing the int8 quantised model which edges at 0.51 loss rate at par with unoptimised float32 model, the quantised model performs much better when deployed on the Sony Spresense. The float32 model carries an inference time of 7.268s per frame which is definitely not suited for real time classification. Comparatively, the int8 model outweighs with 1.544s per frame, 362.5K memory usage and 38.2K Flash.
For the following research, a lot of data analysis and feature engineering has been done for the data ingested into the EdgeImpulse Studio. The following plot is an example of this. While comparing the model performance, this is the relation between the labels of model 1 ( x-axis ) to labels of model 2 ( y-axis ). The linear plot is not completely linear, and hence there is a deviation in results. Data analysis of the segmented LAI helped in finding out model efficacy in this example.
The Leaf Area Index or Biomass for each plant was calculated using image segmentation which was achieved by using the adaptive threshold method for the color information, specifically the Otsu Threshold, followed by a floodfill algorithm in OpenCV and finally using pixel-by-pixel segmented area calculation methods. This pipeline is illustrated below:
The above figure demonstrates the pipeline created by me to process and output Ground Truth LAI using Thresholding method for segmentation. The LAI (Leaf Area Index ) calculated corresponding to the raw image is later used as label for the image and ingested to the EdgeImpulse Studio. The pipeline used in the above process is as follows. An adaptive thresholding mechanism known as Otsu's threshold is used to segment the image from the contrastive background. This is comparatively easy due to the color contrast between the object i.e the plant and the background. However, for instances where the LAI is less than 10cm^2, the Otsu threshold segments the image leaving some noise at the periphery of the confined region. This hampers the overall LAI estimation. Hence, for these images where the plant area is <<< average area, a Floodfill algorithm is used to binarize the noise or holes in image and allow smoother LAI calculation.
This is the defined pipeline for all the samples in the cultivar accumulated, and the process of calculation of LAI for Ground Truth Samples. Post floodfill algorithm being applied, a pixel by pixel area calculation function is applied on binarized images using numpy. The area is calculated in pixels, and using a transformation formula mentioned in the data collection topic, the area in pixels is transformed to LAI.
The formula can be reduced to 2.605 x 10^-2 x Area in pixels cm^2. This formula is only confined when the distance between the lettuce cultivar and camera i.e sony spresense is 78cm.
The final results for Otsu segmentation for all plants in cultivar is as follows -
The Adaptive Thresholding procedure was conducted on a cultivar of Lettuce Flandria plants and used as Ground Truth labels for ingesting the Dataset. A more elaborate view of segmented images per 20 samples is given below:
The python script used for segmentation procedure and adaptive thresholding will be provided in the github repository attached with the code. The analysis of this data in a seaborn plot had been performed above comparing the label numerical value extracted using LAI adaptive thresholding with Day-wise progression. Refer that plot to see complete data analysis of the above segmented pixel by pixel LAI values.
Model Testing and Evaluation:After the model training and data analysis of the segmented labels has been completed, the values predicted from the Regression model trained in EdgeImpulse Studio can be tested to evaluate efficacy on unseen test data. The model performs with near stellar accuracy on testing data evaluation in EdgeImpulse Studio. A test dataset of 19 samples with unique values from 5cm^2 to 90cm^2 LAI is fed to the model. The model evaluates the image data without the input of labels and proposes it's predictions. The predictions within a mean deviation of around 5cm^2. The maximum deviation or error rate withing limited cluster where it's predicted accurate is -4.78. On an average Lettuce Flandria observed much better performance than Lettuce Tiberius. The RMSE calculated for Lettuce Flandria was found out to be 1.185954306 cm^2. The RMSE is calculated using the metrices described below:
The above plot, plotted using seaborn demonstrates a comparative analysis between Ground Truth LAI and RMSE and Predicted LAI and RMSE. The plot explains how the RMSE increases with increase in LAI, and a few anomalies are found in that variation. Overall, its a graph with decreasing slope, and exponentially increasing RMSE with increasing LAI.
The above images are captured from Testing tab in EdgeImpulse Studio. The ingested Test Dataset achieve 100% accuracy which might seem superficial, but it is evaluated on 19 samples with loss of 0.51 which makes is obvious that the model performs exceptionally well! The input Ground Truth Data and predicted Data of LAI by EdgeImpulse Regression model is summarized in the plot following the paragraph:
The plot compares how Predicted LAI performs as compared to Ground Truth LAI for a confined segmented sample. The regression model trained in EI Studio performs and produces accurate predictions for almost all samples. There is an increased error rate in the region between 15-20 cm^2 LAI Ground Truth labels, which indicates the increase in noise in data segmentation in Otsu's segmented images. The Regression model, predicts LAI lower than expected, due to noise in threshold samples, which results in an increase in LAI than expected. The Average RMSE was found to be 1.859 cm^2 which is an indicative factor of accurate predictions. In RMSE, the function, Xo - Xi which is a difference between observed and predicted data. This index was also found out among the data samples predicted and it averaged at -0.2351 cm^2, indicating that the predicted data is on an average 0.235 cm^2 less than Ground Truth.
Above figure represents LAI of segmented images used as test data set to test efficacy. As observed above, the results showed strong correlations between Ground Truth Segmented data and from those predicted by the CNN model. These correlations are measurements concluded from input test images rather than scalar/numerical input data, which makes a huge difference in how effectively CNNs for computer vision models have developed, and become light-weight with increased efficacy without straining on model performance. The quantization of these models done by EdgeImpulse Studio is a massive plus point in Embedded Machine Learning systems most importantly with low power, real time inference in Computer Vision systems.
Deploying Model to Sony Spresense and Real World Data Testing:EdgeImpulse offers a unique compilation system for Embedded ML models which help in quantization of models for upto 55% less RAM! and 35% less ROM! while maintaining consistent accuracy and loss scores. This is a feature I adore about the EdgeImpulse Studio. In the deployment section of EdgeImpulse Studio, there are a list of pre-compiles binaries for supported boards or libraries which can be self-compiled.
For my use case, I'll opt with the Sony Spresense Pre-Compiled Binary which can be directly deployed on the board for real time inference.
With the EON compiler, there is a significant reduction in RAM usage on-board as well as the ROM usage. The RAM usage decreases from 435.6K to 362.5K, nearly 17% reduction in RAM usage, and from 53.5K to 38.2K decrease in ROM/Flash usage, 29% reduction in ROM usage. With the EON Compiler enabled, build the model and flash it over Sony Spresense board. For starters, more info here -
A complete log of compilation and build process can be found at "Build output"
The above figure illustrates flashing the pre-compiled binary to Sony-Spresense board and then next two figure demonstrate real time inference and result estimation on board in under 1s, to be precise, nearly 922ms!
Demonstrating Low Power Consumption and battery operated remote system:
The preceeding images demonstrate the live classification system and real time inference on the Sony Spresense board. The board acquired images from over the plant, inferences the data, processes it and predicts a suitable LAI outcome in real time. The illustrations in images explain the structure of the system, approximate distance and data acquisition procedure for real time on-board inference. The approximate power usage on board is 0.35A per hour which is easily powered by a battery system, here as a powerbank. The tested system lasts for 20.5hours effortlessly over a single charged powerbank. If the clock frequency of the board is set to 32MHz, the average power consumption reduces significantly. The complete system, while in production is expected to be completely battery operated over a suitable voltage power bank, more preferably 1A, which I have used here
The SD card storage on the Sony Spresense can store results of all LAI acquired from over plants in remote laboratories or semi-autonomous/autonomous hydroponic system. The built system is stationary, but a mobile solution can be designed to acquire images corresponding to GPS information tagged with the plant through Sony Spresense. This mobile autonomous system can use and store LAI information per plant collected at specific GPS co-ordinates. There are differentiably plenty applications in the field of auonomous monitoring and growth estimation systems fulfilling UN's SDG's.
All Raw Image Datasets are available at https://figshare.com/s/4e27e3ba666d32daf5c5
All Code and model used for LAI and Biomass estimation is available at - https://github.com/dhruvsheth-ai/Plant-Growth-Estimation-EdgeImpulse
EdgeImpulse Public Dashboard - https://studio.edgeimpulse.com/public/41197/latest
https://studio.edgeimpulse.com/public/47804/latest
Courtesy -
All images and written material/code & algorithms is by Author ( dhruvsheth.linkit@gmail.com ) except those mentioned under [source - xyz]. Model Training and model courtesy - EdgeImpulse, hardware - Sony Spresense [Sony]
You can cite this work -
@misc{Sheth_Plant_Growth_Estimation_2021,
author = {Sheth, Dhruv},
month = {8},
title = {{Plant Growth Estimation using quantised Embedded Regression models for high throughput phenotyping}},
url = {https://github.com/dhruvsheth-ai/Plant-Growth-Estimation-EdgeImpulse/},
year = {2021}
}






_FGZMbuA9tL.jpg)









Comments
Please log in or sign up to comment.