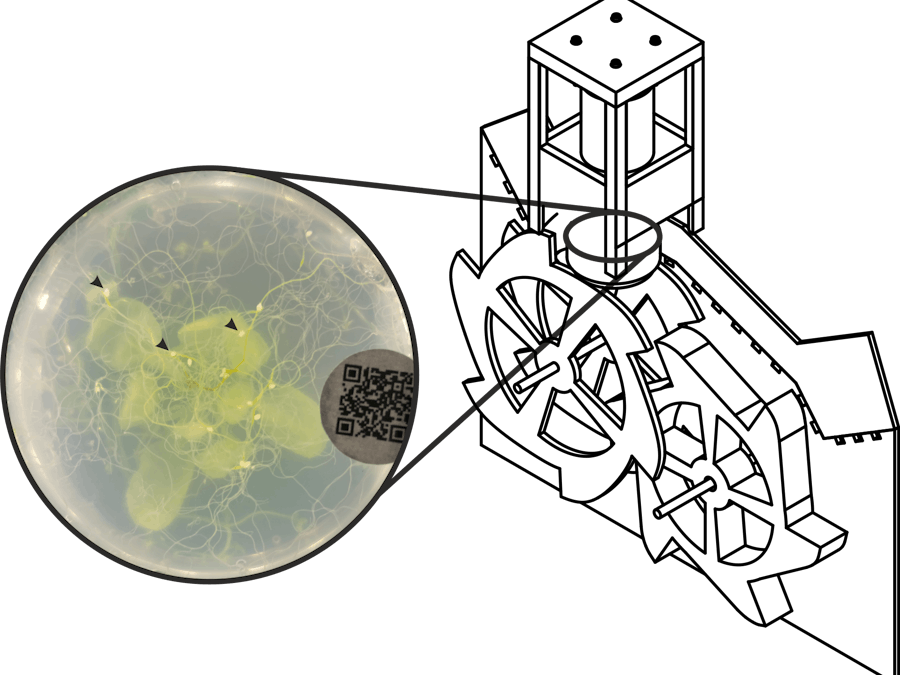
Plant-parasitic nematodes are a major threat to crop production across the world: costing world agriculture $100 billion per year. A substantial barrier to progress in the field is labour intensive phenotyping of these root parasites. To address this constraint, the University of Cambridge and the National Institute of Agricultural Botany (NIAB) are collaborating to automate the phenotyping of plant-nematode infections, using the model plant Arabidopsis thaliana.
To date, we have developed a custom, static, low-cost and easy to use, 3D printed “imaging tower” controlled by a Raspberry Pi computer that can be used to image infected roots of A. thaliana grown in petri dishes. Based on these images, we are developing an analysis platform that combines computer vision and machine learning techniques to recognise, measure and track nematodes from early emergence through to cyst formation. To be able to take full advantage of machine learning phenotyping to screen large populations of plants for nematode resistance, we need a faster way to image plates.
We, therefore, propose to prototype new hardware that would allow low-cost and high-throughput manipulation of thousands of petri dishes in an automated manner. The basic requirements of such a machine would involve developing/scaling up mechanisms for continuous plate de-stacking and stacking, integrating de-stacking and stacking to a single prototype, and integrating our imaging solution into this new hardware between de-stacking and stacking.
Together, this will allow routine phenotyping of mapping-scale populations, accelerate discovery science in this area, and could be more broadly applicable to other systems and entirely unrelated questions based on similar universal petri dish formats.















Comments
Please log in or sign up to comment.